Comparing protein abundance and mRNA expression levels on a genomic scale
- PMID: 12952525
- PMCID: PMC193646
- DOI: 10.1186/gb-2003-4-9-117
Comparing protein abundance and mRNA expression levels on a genomic scale
Abstract
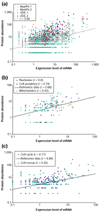
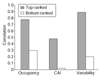
Attempts to correlate protein abundance with mRNA expression levels have had variable success. We review the results of these comparisons, focusing on yeast. In the process, we survey experimental techniques for determining protein abundance, principally two-dimensional gel electrophoresis and mass-spectrometry. We also merge many of the available yeast protein-abundance datasets, using the resulting larger 'meta-dataset' to find correlations between protein and mRNA expression, both globally and within smaller categories.
Figures
References
-
- Klose J. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik. 1975;26:231–243. - PubMed
-
- Hatzimanikatis V, Choe LH, Lee KH. Proteomics: theoretical and experimental considerations. Biotechnol Prog. 1999;15:312–318. - PubMed
-
- Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 1998;16:301–306. - PubMed
-
- McGall GH, Christians FC. High-density genechip oligonucleotide probe arrays. Adv Biochem Eng Biotechnol. 2002;77:21–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

