An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR)
- PMID: 12952533
- PMCID: PMC193654
- DOI: 10.1186/gb-2003-4-9-r54
An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR)
Abstract
Background: Diverse datasets, including genomic, transcriptomic, proteomic and metabolomic data, are becoming readily available for specific organisms. There is currently a need to integrate these datasets within an in silico modeling framework. Constraint-based models of Escherichia coli K-12 MG1655 have been developed and used to study the bacterium's metabolism and phenotypic behavior. The most comprehensive E. coli model to date (E. coli iJE660a GSM) accounts for 660 genes and includes 627 unique biochemical reactions.
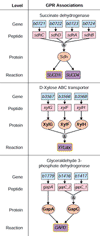
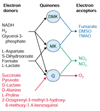
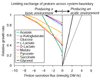
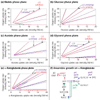
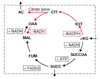
Results: An expanded genome-scale metabolic model of E. coli (iJR904 GSM/GPR) has been reconstructed which includes 904 genes and 931 unique biochemical reactions. The reactions in the expanded model are both elementally and charge balanced. Network gap analysis led to putative assignments for 55 open reading frames (ORFs). Gene to protein to reaction associations (GPR) are now directly included in the model. Comparisons between predictions made by iJR904 and iJE660a models show that they are generally similar but differ under certain circumstances. Analysis of genome-scale proton balancing shows how the flux of protons into and out of the medium is important for maximizing cellular growth.
Conclusions: E. coli iJR904 has improved capabilities over iJE660a. iJR904 is a more complete and chemically accurate description of E. coli metabolism than iJE660a. Perhaps most importantly, iJR904 can be used for analyzing and integrating the diverse datasets. iJR904 will help to outline the genotype-phenotype relationship for E. coli K-12, as it can account for genomic, transcriptomic, proteomic and fluxomic data simultaneously.
Figures

References
-
- Palsson BO. In silico biology through "omics". Nat Biotechnol. 2002;20:649–650. - PubMed
-
- Palsson BO. The challenges of in silico biology. Nat Biotechnol. 2000;18:1147–1150. - PubMed
-
- Edwards JS, Covert M, Palsson B. Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol. 2002;4:133–140. - PubMed
-
- Varma A, Palsson BO. Metabolic flux balancing: basic concepts, scientific and practical use. Bio/Technology. 1994;12:994–998.
-
- Bonarius HPJ, Schmid G, Tramper J. Flux analysis of underdetermined metabolic networks: the quest for the missing constraints. Trends Biotechnol. 1997;15:308–314.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

