Inference of protein function and protein linkages in Mycobacterium tuberculosis based on prokaryotic genome organization: a combined computational approach
- PMID: 12952538
- PMCID: PMC193659
- DOI: 10.1186/gb-2003-4-9-r59
Inference of protein function and protein linkages in Mycobacterium tuberculosis based on prokaryotic genome organization: a combined computational approach
Abstract
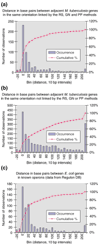
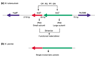
The genome of Mycobacterium tuberculosis was analyzed using recently developed computational approaches to infer protein function and protein linkages. We evaluated and employed a method to infer genes likely to belong to the same operon, as judged by the nucleotide distance between genes in the same genomic orientation, and combined this method with those of the Rosetta Stone, Phylogenetic Profile and conserved Gene Neighbor computational methods for the inference of protein function.
Figures








References
-
- Madigan M, Martinko J, Parker J. Brock Biology of Microorganisms. 9th. New Jersey: Prentice Hall; 2000.
-
- Lodish H, Baltimore D, Berk A, Zipursky SL, Matsudaira P, Darnell J. Molecular Cell Biology. 3rd. New York: Scientific American Books; 1995.
-
- Moreno-Hagelsieb G, Collado-Vides J. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics. 2002;18 Suppl 11:S329–S336. - PubMed
-
- Yada T, Nakao M, Totoki Y, Nakai K. Modeling and predicting transcriptional units of Escherichia coli genes using hidden Markov models. Bioinformatics. 1999;15:987–993. - PubMed

