Assessment of genome-wide protein function classification for Drosophila melanogaster
- PMID: 12952880
- PMCID: PMC403707
- DOI: 10.1101/gr.771603
Assessment of genome-wide protein function classification for Drosophila melanogaster
Abstract
The functional classification of genes on a genome-wide scale is now in its infancy, and we make a first attempt to assess existing methods and identify sources of error. To this end, we compared two independent efforts for associating proteins with functions, one implemented by FlyBase and the other by PANTHER at Celera Genomics. Both methods make inferences based on sequence similarity and the available experimental evidence. However, they differ considerably in methodology and process. Overall, assuming that the systematic error across the two methods is relatively small, we find the protein-to-function association error rate of both the FlyBase and PANTHER methods to be <2%. The primary source of error for both methods appears to be simple human error. Although homology-based inference can certainly cause errors in annotation, our analysis indicates that the frequency of such errors is relatively small compared with the number of correct inferences. Moreover, these homology errors can be minimized by careful tree-based inference, such as that implemented in PANTHER. Often, functional associations are made by one method and not the other, indicating that one of the greatest challenges lies in improving the completeness of available ontology associations.
Figures
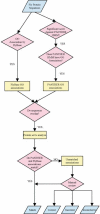

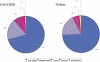



References
-
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. - PubMed
-
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. - PubMed
-
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. - PubMed
WEB SITE REFERENCES
-
- http://panther.celera.com; PANTHER Classification System.
-
- http://www.ensembl.org/Caenorhabditis_briggsae/; Ensembl C. briggsae Genome Server.
-
- http://www.flybase.org; FlyBase@flybase.bio.indiana.edu; FlyBase.
-
- http://www.fruitfly.org/sequence/sequence_db/aa_gadfly.dros.RELEASE2; FlyBase Release 2.
-
- http://www.geneontology.org/; Gene Ontology Consortium.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
