Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells
- PMID: 12952918
- PMCID: PMC182190
- DOI: 10.1172/JCI17464
Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells
Abstract
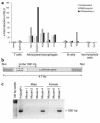
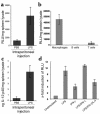
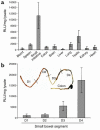
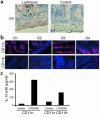
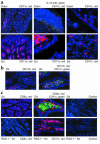
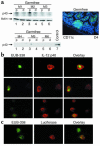
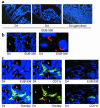
IL-12 p40-related cytokines such as IL-12 p35/p40 heterodimer and IL-23 (p19/p40) are potent regulators of adaptive immune responses. Little is known, however, about the transcriptional regulation of the p40 gene in vivo. In an attempt toward this goal, we have generated transgenic mice expressing firefly luciferase under the control of the IL-12 p40 promoter. High constitutive transgene expression was found in the small intestine only, whereas little reporter gene activity was observed in other tissues. Within the small bowel, constitutive promoter activity was restricted to the terminal ileum and associated with high expression of p40 mRNA as well as p40 and IL-23 p19/p40 proteins. The cells constitutively producing IL-12 p40 were identified as CD8alpha and CD11b double-negative CD11c+ lamina propria dendritic cells (LPDCs) that represent a major cell population in the lamina propria of the small intestine, but not in the colon. FISH directly demonstrated the uptake of bacteria by a subset of LPDCs in the terminal ileum that was associated with p40 expression. Furthermore, little or no p40 protein expression in LPDCs was found in the terminal ileum of germfree mice, indicating a key role of the intestinal flora for constitutive p40 expression. In addition, analysis of transgenic mice with a mutated NF-kappaB target site in the p40 promoter showed a critical role of NF-kappaB for constitutive transgene expression. Our data reveal important functional differences between the mucosal immune systems of the small and large bowel in healthy mice and suggest that the high bacterial load in the terminal ileum activates p40 gene transcription in LPDCs through NF-kappaB. These data suggest a predisposition of the terminal ileum to develop chronic inflammatory responses through IL-23 and thus may provide a molecular explanation for the preferential clinical manifestation of Crohn disease in this part of the gut.
Figures

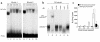

Comment in
-
Dendritic cells and the intestinal bacterial flora: a role for localized mucosal immune responses.J Clin Invest. 2003 Sep;112(5):648-51. doi: 10.1172/JCI19545. J Clin Invest. 2003. PMID: 12952911 Free PMC article.
References
-
- Wolf SF, et al. Cloning of cDNA for natural killer cell stimulatory factor, a heterodimeric cytokine with multiple biologic effects on T and natural killer cells. J. Immunol. 1991;146:3074–3081. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

