Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei
- PMID: 12952935
- PMCID: PMC2172812
- DOI: 10.1083/jcb.200304096
Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei
Abstract
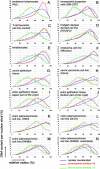
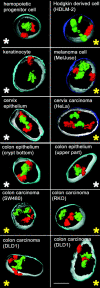
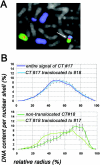
A gene density-related difference in the radial arrangement of chromosome territories (CTs) was previously described for human lymphocyte nuclei with gene-poor CT #18 located toward the nuclear periphery and gene-dense CT #19 in the nuclear interior (Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. J. Cell Biol. 145:1119-1131). Here, we analyzed the radial distribution of chromosome 18 and 19 chromatin in six normal cell types and in eight tumor cell lines, some of them with imbalances and rearrangements of the two chromosomes. Our findings demonstrate that a significant difference in the radial distribution of #18 and #19 chromatin is a common feature of higher order chromatin architecture in both normal and malignant cell types. However, in seven of eight tumor cell lines, the difference was less pronounced compared with normal cell nuclei due to a higher fraction of nuclei showing an inverted CT position, i.e., a CT #18 located more internally than a CT #19. This observation emphasizes a partial loss of radial chromatin order in tumor cell nuclei.
Figures


References
-
- Abdel-Rahman, W.M., K. Katsura, W. Rens, P.A. Gorman, D. Sheer, D. Bicknell, W.F. Bodmer, M.J. Arends, A.H. Wyllie, and P.A. Edwards. 2001. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl. Acad. Sci. USA. 98:2538–2543. - PMC - PubMed
-
- Bach, S.P., A.G. Renehan, and C.S. Potten. 2000. Stem cells: the intestinal stem cell as a paradigm. Carcinogenesis. 21:469–476. - PubMed
-
- Bauer, R.K. 1962. Der median-quartile-test. Metrika. 5:1–16.
-
- Bennett, D.C., K. Bridges, and I.A. McKay. 1985. Clonal separation of mature melanocytes from premelanocytes in a diploid human cell strain: spontaneous and induced pigmentation of premelanocytes. J. Cell Sci. 77:167–183. - PubMed
-
- Boyle, S., S. Gilchrist, J.M. Bridger, N.L. Mahy, J.A. Ellis, and W.A. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

