Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment
- PMID: 12953091
- PMCID: PMC2194186
- DOI: 10.1084/jem.20030846
Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment
Abstract
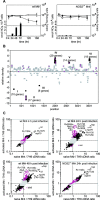
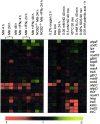
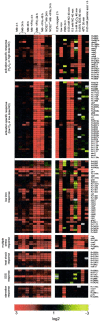
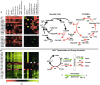
Little is known about the biochemical environment in phagosomes harboring an infectious agent. To assess the state of this organelle we captured the transcriptional responses of Mycobacterium tuberculosis (MTB) in macrophages from wild-type and nitric oxide (NO) synthase 2-deficient mice before and after immunologic activation. The intraphagosomal transcriptome was compared with the transcriptome of MTB in standard broth culture and during growth in diverse conditions designed to simulate features of the phagosomal environment. Genes expressed differentially as a consequence of intraphagosomal residence included an interferon gamma- and NO-induced response that intensifies an iron-scavenging program, converts the microbe from aerobic to anaerobic respiration, and induces a dormancy regulon. Induction of genes involved in the activation and beta-oxidation of fatty acids indicated that fatty acids furnish carbon and energy. Induction of sigmaE-dependent, sodium dodecyl sulfate-regulated genes and genes involved in mycolic acid modification pointed to damage and repair of the cell envelope. Sentinel genes within the intraphagosomal transcriptome were induced similarly by MTB in the lungs of mice. The microbial transcriptome thus served as a bioprobe of the MTB phagosomal environment, showing it to be nitrosative, oxidative, functionally hypoxic, carbohydrate poor, and capable of perturbing the pathogen's cell envelope.
Figures

References
-
- Aderem, A., and D.M. Underhill. 1999. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 17:593–623. - PubMed
-
- Amer, A.O., and M.S. Swanson. 2002. A phagosome of one's own: a microbial guide to life in the macrophage. Curr. Opin. Microbiol. 5:56–61. - PubMed
-
- Russell, D.G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2:569–577. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

