Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program
- PMID: 12953092
- PMCID: PMC2194188
- DOI: 10.1084/jem.20030205
Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program
Abstract
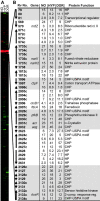
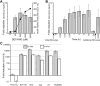
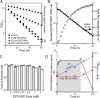
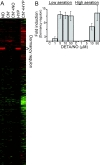
An estimated two billion persons are latently infected with Mycobacterium tuberculosis. The host factors that initiate and maintain this latent state and the mechanisms by which M. tuberculosis survives within latent lesions are compelling but unanswered questions. One such host factor may be nitric oxide (NO), a product of activated macrophages that exhibits antimycobacterial properties. Evidence for the possible significance of NO comes from murine models of tuberculosis showing progressive infection in animals unable to produce the inducible isoform of NO synthase and in animals treated with a NO synthase inhibitor. Here, we show that O2 and low, nontoxic concentrations of NO competitively modulate the expression of a 48-gene regulon, which is expressed in vivo and prepares bacilli for survival during long periods of in vitro dormancy. NO was found to reversibly inhibit aerobic respiration and growth. A heme-containing enzyme, possibly the terminal oxidase in the respiratory pathway, likely senses and integrates NO and O2 levels and signals the regulon. These data lead to a model postulating that, within granulomas, inhibition of respiration by NO production and O2 limitation constrains M. tuberculosis replication rates in persons with latent tuberculosis.
Figures

References
-
- Dye, C., S. Scheele, P. Dolin, V. Pathania, and M.C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 282:677–686. - PubMed
-
- McKinney, J.D., K. Honer zu Bentrup, E.J. Munoz-Elias, A. Miczak, B. Chen, W.T. Chan, D. Swenson, J.C. Sacchettini, W.R. Jacobs, Jr., and D.G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 406:735–738. - PubMed
-
- Betts, J.C., P.T. Lukey, L.C. Robb, R.A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731. - PubMed
-
- Wayne, L.G., and C.D. Sohaskey. 2001. Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

