A grape ASR protein involved in sugar and abscisic acid signaling
- PMID: 12953118
- PMCID: PMC181338
- DOI: 10.1105/tpc.013854
A grape ASR protein involved in sugar and abscisic acid signaling
Abstract
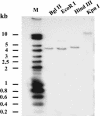
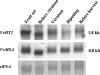
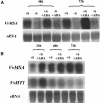
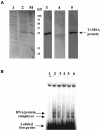
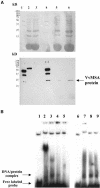
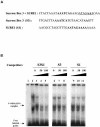
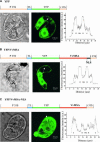
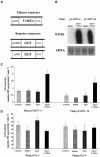
The function of ASR (ABA [abscisic acid]-, stress-, and ripening-induced) proteins remains unknown. A grape ASR, VvMSA, was isolated by means of a yeast one-hybrid approach using as a target the proximal promoter of a grape putative monosaccharide transporter (VvHT1). This promoter contains two sugar boxes, and its activity is induced by sucrose and glucose. VvMSA and VvHT1 share similar patterns of expression during the ripening of grape. Both genes are inducible by sucrose in grape berry cell culture, and sugar induction of VvMSA is enhanced strongly by ABA. These data suggest that VvMSA is involved in a common transduction pathway of sugar and ABA signaling. Gel-shift assays demonstrate a specific binding of VvMSA to the 160-bp fragment of the VvHT1 promoter and more precisely to two sugar-responsive elements present in this target. The positive regulation of VvHT1 promoter activity by VvMSA also is shown in planta by coexpression experiments. The nuclear localization of the yellow fluorescent protein-VvMSA fusion protein and the functionality of the VvMSA nuclear localization signal are demonstrated. Thus, a biological function is ascribed to an ASR protein. VvMSA acts as part of a transcription-regulating complex involved in sugar and ABA signaling.
Figures


Similar articles
-
Grape ASR Regulates Glucose Transport, Metabolism and Signaling.Int J Mol Sci. 2022 May 31;23(11):6194. doi: 10.3390/ijms23116194. Int J Mol Sci. 2022. PMID: 35682874 Free PMC article.
-
Cloning and expression of a hexose transporter gene expressed during the ripening of grape berry.Plant Physiol. 1999 Aug;120(4):1083-94. doi: 10.1104/pp.120.4.1083. Plant Physiol. 1999. PMID: 10444092 Free PMC article.
-
Grape ASR-Silencing Sways Nuclear Proteome, Histone Marks and Interplay of Intrinsically Disordered Proteins.Int J Mol Sci. 2022 Jan 28;23(3):1537. doi: 10.3390/ijms23031537. Int J Mol Sci. 2022. PMID: 35163458 Free PMC article.
-
Sugar-regulated expression of a putative hexose transport gene in grape.Plant Physiol. 2003 Jan;131(1):326-34. doi: 10.1104/pp.009522. Plant Physiol. 2003. PMID: 12529540 Free PMC article.
-
A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter.Plant Cell. 2003 Sep;15(9):2076-92. doi: 10.1105/tpc.014597. Plant Cell. 2003. PMID: 12953112 Free PMC article.
Cited by
-
Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato.Plant Physiol. 2012 Jan;158(1):283-98. doi: 10.1104/pp.111.186866. Epub 2011 Nov 22. Plant Physiol. 2012. PMID: 22108525 Free PMC article.
-
An ABA-responsive element in the AtSUC1 promoter is involved in the regulation of AtSUC1 expression.Planta. 2010 Sep;232(4):911-23. doi: 10.1007/s00425-010-1228-4. Epub 2010 Jul 16. Planta. 2010. PMID: 20635094
-
Gene expression analyses in individual grape (Vitis vinifera L.) berries during ripening initiation reveal that pigmentation intensity is a valid indicator of developmental staging within the cluster.Plant Mol Biol. 2008 Oct;68(3):301-15. doi: 10.1007/s11103-008-9371-z. Epub 2008 Jul 19. Plant Mol Biol. 2008. PMID: 18642093
-
Comparison of Transcriptional Response of C3 and C4 Plants to Drought Stress Using Meta-Analysis and Systems Biology Approach.Front Plant Sci. 2021 Jul 1;12:668736. doi: 10.3389/fpls.2021.668736. eCollection 2021. Front Plant Sci. 2021. PMID: 34276729 Free PMC article.
-
TaASR1-D confers abiotic stress resistance by affecting ROS accumulation and ABA signalling in transgenic wheat.Plant Biotechnol J. 2021 Aug;19(8):1588-1601. doi: 10.1111/pbi.13572. Epub 2021 Mar 23. Plant Biotechnol J. 2021. PMID: 33638922 Free PMC article.
References
-
- Bernard, M., Dinet, V., and Voisin, P. (2001). Transcriptional regulation of the chicken hydroxyindole-O-methyltransferase gene by the cone-rod homeobox-containing protein. J. Neurochem. 79, 248–257. - PubMed
-
- Blouin, J., and Guimberteau, G. (2000). Maturité. In Maturation et Maturité des Raisins. (Bordeaux, France: Féret), pp. 29–31.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

