The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1
- PMID: 12953119
- PMCID: PMC181339
- DOI: 10.1105/tpc.012849
The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1
Abstract
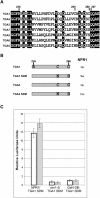
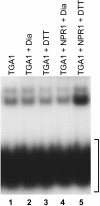
The Arabidopsis NPR1 protein is essential for regulating salicylic acid-dependent gene expression during systemic acquired resistance. NPR1 interacts differentially with members of the TGA class of basic domain/Leu zipper transcription factors and regulates their DNA binding activity. Here, we report that although TGA1 does not interact with NPR1 in yeast two-hybrid assays, treatment with salicylic acid induces the interaction between these proteins in Arabidopsis leaves. This phenomenon is correlated with a reduction of TGA1 Cys residues. Furthermore, site-directed mutagenesis of TGA1 Cys-260 and Cys-266 enables the interaction with NPR1 in yeast and Arabidopsis. Together, these results indicate that TGA1 relies on the oxidation state of Cys residues to mediate the interaction with NPR1. An intramolecular disulfide bridge in TGA1 precludes interaction with NPR1, and NPR1 can only stimulate the DNA binding activity of the reduced form of TGA1. Unlike its animal and yeast counterparts, the DNA binding activity of TGA1 is not redox regulated; however, this property is conferred by interaction with the NPR1 cofactor.
Figures




References
-
- Abate, C., Patel, L., Rauscher, F.J., III, and Curran, T. (1990). Redox regulation of fos and jun DNA-binding activity in vitro. Science 249, 1157–1161. - PubMed
-
- Alvarez, M.E., Pennell, R.I., Meijer, P.J., Ishikawa, A., Dixon, R.A., and Lamb, C. (1998). Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92, 773–784. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. - PubMed
-
- Bayer, E.A., Safars, M., and Wilchek, M. (1987). Selective labeling of sulfhydryls and disulfides on blot transfers using avidin-biotin technology: Studies on purified proteins and erythrocyte membranes. Anal. Biochem. 161, 262–271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

