Differential regulation of the glucose-6-phosphatase TATA box by intestine-specific homeodomain proteins CDX1 and CDX2
- PMID: 12954759
- PMCID: PMC203330
- DOI: 10.1093/nar/gkg747
Differential regulation of the glucose-6-phosphatase TATA box by intestine-specific homeodomain proteins CDX1 and CDX2
Abstract
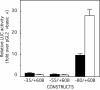
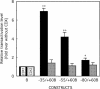
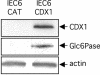
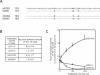
Glucose-6-phosphatase (Glc6Pase), the last enzyme of gluconeogenesis, is only expressed in the liver, kidney and small intestine. The expression of the Glc6Pase gene exhibits marked specificities in the three tissues in various situations, but the molecular basis of the tissue specificity is not known. The presence of a consensus binding site of CDX proteins in the minimal Glc6Pase gene promoter has led us to consider the hypothesis that these intestine-specific CDX factors could be involved in the Glc6Pase-specific expression in the small intestine. We first show that the Glc6Pase promoter is active in both hepatic HepG2 and intestinal CaCo2 cells. Using gel shift mobility assay, mutagenesis and competition experiments, we show that both CDX1 and CDX2 can bind the minimal promoter, but only CDX1 can transactivate it. Consistently, intestinal IEC6 cells stably overexpressing CDX1 exhibit induced expression of the Glc6Pase protein. We demonstrate that a TATAAAA sequence, located in position -31/-25 relating to the transcription start site, exhibits separable functions in the preinitiation of transcription and the transactivation by CDX1. Disruption of this site dramatically suppresses both basal transcription and the CDX1 effect. The latter may be restored by inserting a couple of CDX- binding sites in opposite orientation similar to that found in the sucrase-isomaltase promoter. We also report that the specific stimulatory effect of CDX1 on the Glc6Pase TATA-box, compared to CDX2, is related to the fact that CDX1, but not CDX2, can interact with the TATA-binding protein. Together, these data strongly suggest that CDX proteins could play a crucial role in the specific expression of the Glc6Pase gene in the small intestine. They also suggest that CDX transactivation might be essential for intestine gene expression, irrespective of the presence of a functional TATA box.
Figures


Similar articles
-
Repression of the desmocollin 2 gene expression in human colon cancer cells is relieved by the homeodomain transcription factors Cdx1 and Cdx2.Mol Cancer Res. 2008 Sep;6(9):1478-90. doi: 10.1158/1541-7786.MCR-07-2161. Mol Cancer Res. 2008. PMID: 18819935
-
Differential regulation of intestinal alkaline phosphatase gene expression by Cdx1 and Cdx2.Am J Physiol Gastrointest Liver Physiol. 2005 Aug;289(2):G285-90. doi: 10.1152/ajpgi.00037.2005. Epub 2005 Mar 17. Am J Physiol Gastrointest Liver Physiol. 2005. PMID: 15774940
-
cis-Acting elements and transcription factors involved in the intestinal specific expression of the rat calbindin-D9K gene: binding of the intestine-specific transcription factor Cdx-2 to the TATA box.Eur J Biochem. 1996 Mar 15;236(3):778-88. doi: 10.1111/j.1432-1033.1996.00778.x. Eur J Biochem. 1996. PMID: 8665895
-
The role of Cdx proteins in intestinal development and cancer.Cancer Biol Ther. 2004 Jul;3(7):593-601. doi: 10.4161/cbt.3.7.913. Epub 2004 Jul 9. Cancer Biol Ther. 2004. PMID: 15136761 Review.
-
CDX-2, a new marker for adenocarcinoma of gastrointestinal origin.Adv Anat Pathol. 2004 Mar;11(2):101-5. doi: 10.1097/00125480-200403000-00004. Adv Anat Pathol. 2004. PMID: 15090846 Review.
Cited by
-
Friend of GATA suppresses the GATA-induced transcription of hepcidin in hepatocytes through a GATA-regulatory element in the HAMP promoter.J Mol Endocrinol. 2011 Nov 21;47(3):299-313. doi: 10.1530/JME-11-0060. Print 2011 Dec. J Mol Endocrinol. 2011. PMID: 21971825 Free PMC article.
-
Huntingtin-associated protein 1: Eutherian adaptation from a TRAK-like protein, conserved gene promoter elements, and localization in the human intestine.BMC Evol Biol. 2016 Oct 13;16(1):214. doi: 10.1186/s12862-016-0780-3. BMC Evol Biol. 2016. PMID: 27737633 Free PMC article.
-
The suppression of hepatic glucose production improves metabolism and insulin sensitivity in subcutaneous adipose tissue in mice.Diabetologia. 2016 Dec;59(12):2645-2653. doi: 10.1007/s00125-016-4097-y. Epub 2016 Sep 9. Diabetologia. 2016. PMID: 27631137
-
Glucotoxicity induces glucose-6-phosphatase catalytic unit expression by acting on the interaction of HIF-1α with CREB-binding protein.Diabetes. 2012 Oct;61(10):2451-60. doi: 10.2337/db11-0986. Epub 2012 Jul 10. Diabetes. 2012. PMID: 22787137 Free PMC article.
-
Intestinal mucosal atrophy and adaptation.World J Gastroenterol. 2012 Nov 28;18(44):6357-75. doi: 10.3748/wjg.v18.i44.6357. World J Gastroenterol. 2012. PMID: 23197881 Free PMC article. Review.
References
-
- Mithieux G. (1997) New knowledge regarding glucose-6 phosphatase gene and protein and their roles in the regulation of glucose metabolism. Eur. J. Endocrinol., 136, 137–145. - PubMed
-
- Croset M., Rajas,F., Zitoun,C., Hurot,J.M., Montano,S. and Mithieux,G. (2001) Rat small intestine is an insulin-sensitive gluconeogenic organ. Diabetes, 50, 740–746. - PubMed
-
- Mithieux G. (2001) New data and concepts on glutamine and glucose metabolism in the gut. Curr. Opin. Clin. Nutr. Metab. Care, 4, 267–271. - PubMed
-
- Chatelain F., Pegorier,J.P., Minassian,C., Bruni,N., Tarpin,S., Girard,J. and Mithieux,G. (1998) Development and regulation of glucose-6-phosphatase gene expression in rat liver, intestine and kidney: in vivo and in vitro studies in cultured fetal hepatocytes. Diabetes, 47, 882–889. - PubMed
-
- Rajas F., Bruni,N., Montano,S., Zitoun,C. and Mithieux,G. (1999) The glucose-6 phosphatase gene is expressed in human and rat small intestine: regulation of expression in fasted and diabetic rats. Gastroenterology, 117, 132–139. - PubMed

