Light-mediated regulation defines a minimal promoter region of TOP2
- PMID: 12954761
- PMCID: PMC203327
- DOI: 10.1093/nar/gkg744
Light-mediated regulation defines a minimal promoter region of TOP2
Abstract
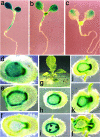
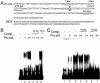
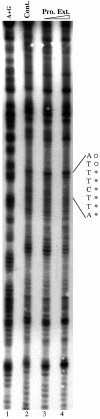
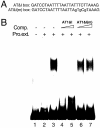
Light signaling has been demonstrated to be an important factor for plant growth and development; however, its role in the regulation of DNA replication and cell cycle has just started to be unraveled. In this work, we have demonstrated that the TOP2 promoter of Pisum sativum (pea) is activated by a broad spectrum of light including far-red light (FR), red light (RL) and blue light (BL). Deletion analyses of the TOP2 promoter in transformed plants, Arabidopsis thaliana and Nicotiana tobaccum (tobacco), define a minimal promoter region that is induced by RL, FR and BL, and is essential and sufficient for light-mediated activation. The minimal promoter of TOP2 follows the phytochrome- mediated low-fluence response similar to complex light regulated promoters. DNA-protein interaction studies reveal the presence of a DNA binding activity specific to a 106 bp region of the minimal promoter that is crucial for light-mediated activation. These results altogether indicate a direct involvement of light signaling in the regulation of expression of TOP2, one of the components of the DNA replication/cell cycle machinery.
Figures
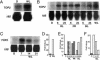

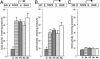

References
-
- Champoux J.J. (1990) Mechanistic aspect of type I topoisomerases. In Cozzarelli,N.R. and Wang,J.C. (eds), DNA Topology and its Biological Effects. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 217–242.
-
- Hsieh T.S. (1990) Mechanistic aspects of type II topoisomerases. In Cozzarelli,N.R. and Wang,J.C. (eds), DNA Topology and its Biological Effects. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 243–263.
-
- Wang J.C. (1996) DNA topoisomerases. Annu. Rev. Biochem., 65, 635–692. - PubMed
-
- Madhusudan K. and Nagaraja,V. (1996) Alignment and phylogenetic analysis of type II topoisomerases. Alignment and phylogenetic analysis of type II topoisomerases. J. Biosci., 21, 613–629.
-
- Wang J.C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev., 3, 430–440. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

