Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein
- PMID: 12954769
- PMCID: PMC203308
- DOI: 10.1093/nar/gkg715
Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein
Abstract
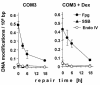
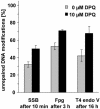
Activation of poly(ADP-ribose)polymerases 1 and 2 (PARP-1 and PARP-2) is one of the earliest responses of mammalian cells to DNA damage by numerous genotoxic agents. We have analysed the influence of PARP inhibition, either achieved by over-expression of the DNA binding domain of PARP-1 or by treatment with 3,4-dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone, on the repair of single-strand breaks (SSB), pyrimidine dimers and oxidative base modifications sensitive to Fpg protein (mostly 8-hydroxyguanine) in mammalian cells at very low, non-cytotoxic levels of DNA damage. The data show that the repair rates of all three types of DNA damage are significantly lower in PARP-inhibited cells. Importantly, the retardation of the repair of base modifications is not associated with accumulation of intermediates such as SSB or abasic sites. Moreover, the influence of the PARP inhibition is not observed in cells deficient in Cockayne syndrome B protein (Csb). The results indicate that PARP activation and Csb are both involved in a novel mechanism that accelerates the global repair of various types of DNA modifications.
Figures



References
-
- Shall S. and de Murcia,G. (2000) Poly(ADP-ribose)polymerase-1: what have we learned from the deficient mouse model? Mutat. Res., 460, 1–15. - PubMed
-
- Bürkle A. (2001) Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett., 163, 1–5. - PubMed
-
- Bürkle A. (2001) Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays, 23, 795–806. - PubMed
-
- Herceg Z. and Wang,Z.Q. (2001) Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res., 477, 97–110. - PubMed
-
- Ziegler M. and Oei,S.L. (2001) A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays, 23, 543–548. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

