Identification of Daxx interacting with p73, one of the p53 family, and its regulation of p53 activity by competitive interaction with PML
- PMID: 12954772
- PMCID: PMC203324
- DOI: 10.1093/nar/gkg741
Identification of Daxx interacting with p73, one of the p53 family, and its regulation of p53 activity by competitive interaction with PML
Abstract
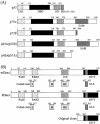
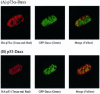
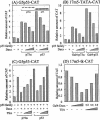
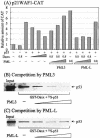
We performed a yeast two-hybrid screen using p73alpha, which is a member of the p53 family, as bait. We found that the p53 family members were functionally associated with Daxx, which was described originally as a cytoplasmic mediator of Fas signaling, but has been identified recently as a nuclear protein that co-localizes with the promyelocytic leukemia (PML) protein and regulates transcription. Extensive yeast two-hybrid assays indicated a physical interaction between a region including the oligomerization domain (OD) of p73alpha (amino acids 345-380) or p53 (amino acids 319-360) and amino acids 161-311 and 667-740 (C-terminal S/P/T-rich domain) of hDaxx, which is the common binding region of Fas, ASK1 and PML. This interaction was further confirmed by in vitro GST pull-down and in vivo immunoprecipitation assays. Both Daxx and p73/p53 co-localized in nuclear dot-like structures, which are probably nuclear PML oncogenic domains (PODs) or the nuclear domain NB10. Transient co-expression of Daxx resulted in strong inhibition of p73- and p53-mediated transcriptional activation of the synthetic p53-responsive and p21WAF1 promoters. Consequently, Gal4-Daxx repressed basal transcription in a dose-dependent manner. Treatment with trichostatin A, which is an inhibitor of histone deacetylase, or PML over-expression relieved Daxx-mediated transcriptional repression of p53. The mechanism underlying PML-mediated derepression appears to be competitive binding between Daxx, p53 and PML. Taken together, these findings delineate a transcriptional regulatory network that is modulated by differential Daxx-p53-PML interactions in the nuclear PODs. Therefore, Daxx is implicated in the regulation of the cell cycle and apoptosis through transcriptional regulation of p53 and possibly its family members.
Figures






Similar articles
-
Identification and characterization of HIPK2 interacting with p73 and modulating functions of the p53 family in vivo.J Biol Chem. 2002 Aug 30;277(35):32020-8. doi: 10.1074/jbc.M200153200. Epub 2002 Mar 29. J Biol Chem. 2002. PMID: 11925430
-
Adenovirus E1B 55-kilodalton oncoprotein binds to Daxx and eliminates enhancement of p53-dependent transcription by Daxx.J Virol. 2003 Nov;77(21):11809-21. doi: 10.1128/jvi.77.21.11809-11821.2003. J Virol. 2003. PMID: 14557665 Free PMC article.
-
Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs).EMBO J. 1999 Nov 1;18(21):6037-49. doi: 10.1093/emboj/18.21.6037. EMBO J. 1999. PMID: 10545115 Free PMC article.
-
The role of PML in tumor suppression.Cell. 2002 Jan 25;108(2):165-70. doi: 10.1016/s0092-8674(02)00626-8. Cell. 2002. PMID: 11832207 Review.
-
PML interaction with p53 and its role in apoptosis and replicative senescence.Oncogene. 2001 Oct 29;20(49):7250-6. doi: 10.1038/sj.onc.1204856. Oncogene. 2001. PMID: 11704853 Review.
Cited by
-
DNA damage-induced regulatory interplay between DAXX, p53, ATM kinase and Wip1 phosphatase.Cell Cycle. 2015;14(3):375-87. doi: 10.4161/15384101.2014.988019. Cell Cycle. 2015. PMID: 25659035 Free PMC article.
-
Reciprocal regulation of Daxx and PIK3CA promotes colorectal cancer cell growth.Cell Mol Life Sci. 2022 Jun 19;79(7):367. doi: 10.1007/s00018-022-04399-8. Cell Mol Life Sci. 2022. PMID: 35718818 Free PMC article.
-
Daxx-beta and Daxx-gamma, two novel splice variants of the transcriptional co-repressor Daxx.J Biol Chem. 2011 Jun 3;286(22):19576-88. doi: 10.1074/jbc.M110.196311. Epub 2011 Apr 10. J Biol Chem. 2011. PMID: 21482821 Free PMC article.
-
NSP 5a3a: a potential novel cancer target in head and neck carcinoma.Oncotarget. 2010 Oct;1(6):423-435. doi: 10.18632/oncotarget.176. Oncotarget. 2010. PMID: 21311098 Free PMC article.
-
Daxx is a transcriptional repressor of CCAAT/enhancer-binding protein beta.J Biol Chem. 2009 Oct 16;284(42):28783-94. doi: 10.1074/jbc.M109.041186. Epub 2009 Aug 18. J Biol Chem. 2009. PMID: 19690170 Free PMC article.
References
-
- Somasundaram K. and El-Deiry,W.S. (2000) Tumor suppressor p53: regulation and function. Front. Biosci., 5, 424–437. - PubMed
-
- Sang B.C., Chen,J.Y., Minna,J. and Barbosa,M.S. (1994) Distinct regions of p53 have a differential role in transcriptional activation and repression functions. Oncogene, 9, 853–859. - PubMed
-
- Shaulian E., Haviv,I., Shaul,Y. and Oren,M. (1995) Transcriptional repression by the C-terminal domain of p53. Oncogene, 10, 671–680. - PubMed
-
- Hong T.M., Chen,J.J., Peck,K., Yang,P.C. and Wu,C.W. (2001) p53 amino acids 339–346 represent the minimal p53 repression domain. J. Biol. Chem., 276, 1510–1515. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

