Rapid generation of gene disruption constructs by in vitro transposition and identification of a Dictyostelium protein kinase that regulates its rate of growth and development
- PMID: 12954783
- PMCID: PMC203333
- DOI: 10.1093/nar/gng095
Rapid generation of gene disruption constructs by in vitro transposition and identification of a Dictyostelium protein kinase that regulates its rate of growth and development
Abstract
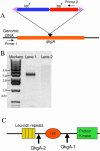
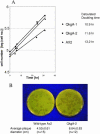
We describe a rapid method for creating Dictyo stelium gene disruption constructs, whereby the target gene is interrupted by a drug resistance cassette using in vitro transposition. A fragment of genomic DNA containing the gene to be disrupted is amplified by PCR, cloned into a plasmid vector using topoisomerase and then employed as the substrate in an in vitro Tn5 transposition reaction. The transposing species is a fragment of DNA containing a Dictyostelium blasticidin S resistance (bs(r)) cassette linked to a bacterial tetracycline resistance (tet(r)) cassette. After transposition the plasmid DNA is transformed into Escherichia coli and clones in which the bs(r)-tet(r) cassette is inserted into the Dictyostelium target DNA are identified. To demonstrate its utility we have employed the method to disrupt the gene encoding QkgA, a novel protein kinase identified from the Dictyostelium genome sequencing project. QkgA is structurally homologous to two previously identified Dictyostelium kinases, GbpC and pats1. Like them it contains a leucine-rich repeat domain, a small GTP-binding (ras) domain and a MEKK domain. Disruption of the qkgA gene causes a marked increase in growth rate and, during development, aggregation occurs relatively slowly to form abnormally large multicellular structures.
Figures




References
-
- Hamer L., DeZwaan,T.M., Montenegro-Chamorro,M.V., Frank,S.A. and Hamer,J.E. (2001) Recent advances in large-scale transposon mutagenesis. Curr. Opin. Chem. Biol., 5, 67–73. - PubMed
-
- Kessin R.H. (2001) Dictyostelium—Evolution, Cell Biology and the Development of Multicellularity. Cambridge University Press, Cambridge, UK.
-
- Glockner G., Eichinger,L., Szafranski,K., Pachebat,J.A., Bankier,A.T., Dear,P.H., Lehmann,D., Baumgart,C., Parra,G., Abril,J.F. et al. (2002) Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature, 418, 79–85. - PubMed
-
- Morio T., Urushihara,H., Saito,T., Ugawa,Y., Mizuno,H., Yoshida,M., Yoshino,R., Mitra,B.N., Pi,M., Sato,T. et al. (1998) The Dictyostelium developmental cDNA project: generation and analysis of expressed sequence tags from the first-finger stage of development. DNA Res., 5, 335–340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

