Morphological, host range, and genetic characterization of two coliphages
- PMID: 12957924
- PMCID: PMC194992
- DOI: 10.1128/AEM.69.9.5364-5371.2003
Morphological, host range, and genetic characterization of two coliphages
Abstract
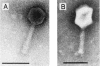
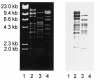
Two coliphages, AR1 and LG1, were characterized based on their morphological, host range, and genetic properties. Transmission electron microscopy showed that both phages belonged to the Myoviridae; phage particles of LG1 were smaller than those of AR1 and had an isometric head 68 nm in diameter and a complex contractile tail 111 nm in length. Transmission electron micrographs of AR1 showed phage particles consisting of an elongated isometric head of 103 by 74 nm and a complex contractile tail 116 nm in length. Both phages were extensively tested on many strains of Escherichia coli and other enterobacteria. The results showed that both phages could infect many serotypes of E. coli. Among the enterobacteria, Proteus mirabilis, Shigella dysenteriae, and two Salmonella strains were lysed by the phages. The genetic material of AR1 and LG1 was characterized. Phage LG1 had a genome size of 49.5 kb compared to 150 kb for AR1. Restriction endonuclease analysis showed that several restriction enzymes could degrade DNA from both phages. The morphological, genome size, and restriction endonuclease similarities between AR1 and phage T4 were striking. Southern hybridizations showed that AR1 and T4 are genetically related. The wide host ranges of phages AR1 and LG1 suggest that they may be useful as biocontrol, therapeutic, or diagnostic agents to control and detect the prevalence of E. coli in animals and food.
Figures
Similar articles
-
Isolation and Characterization of Lytic Phage vB_EcoM_JS09 against Clinically Isolated Antibiotic-Resistant Avian Pathogenic Escherichia coli and Enterotoxigenic Escherichia coli.Intervirology. 2015;58(4):218-31. doi: 10.1159/000437426. Epub 2015 Aug 26. Intervirology. 2015. PMID: 26337345
-
Isolation and characterization of a T4-like phage with a relatively wide host range within Escherichia coli.J Basic Microbiol. 2016 Apr;56(4):405-21. doi: 10.1002/jobm.201500440. Epub 2015 Dec 23. J Basic Microbiol. 2016. PMID: 26697952
-
Identification and characterization of new broad host-range rV5-like coliphages C203 and P206 directed against enterobacteria.Infect Genet Evol. 2018 Oct;64:254-261. doi: 10.1016/j.meegid.2018.07.004. Epub 2018 Jul 4. Infect Genet Evol. 2018. PMID: 30033383
-
T4-like phage Bp7, a potential antimicrobial agent for controlling drug-resistant Escherichia coli in chickens.Appl Environ Microbiol. 2013 Sep;79(18):5559-65. doi: 10.1128/AEM.01505-13. Epub 2013 Jul 8. Appl Environ Microbiol. 2013. PMID: 23835183 Free PMC article.
-
The SPO1-related bacteriophages.Arch Virol. 2010 Oct;155(10):1547-61. doi: 10.1007/s00705-010-0783-0. Epub 2010 Aug 17. Arch Virol. 2010. PMID: 20714761 Review.
Cited by
-
T4-like Bacteriophages Isolated from Pig Stools Infect Yersinia pseudotuberculosis and Yersinia pestis Using LPS and OmpF as Receptors.Viruses. 2021 Feb 13;13(2):296. doi: 10.3390/v13020296. Viruses. 2021. PMID: 33668618 Free PMC article.
-
Coliphages as viral indicators of sanitary significance for drinking water.Front Microbiol. 2022 Jul 26;13:941532. doi: 10.3389/fmicb.2022.941532. eCollection 2022. Front Microbiol. 2022. PMID: 35958148 Free PMC article. Review.
-
Isolation and Characterization of phiLLS, a Novel Phage with Potential Biocontrol Agent against Multidrug-Resistant Escherichia coli.Front Microbiol. 2017 Jul 21;8:1355. doi: 10.3389/fmicb.2017.01355. eCollection 2017. Front Microbiol. 2017. PMID: 28785246 Free PMC article.
-
Guidelines to Compose an Ideal Bacteriophage Cocktail.Methods Mol Biol. 2024;2734:49-66. doi: 10.1007/978-1-0716-3523-0_4. Methods Mol Biol. 2024. PMID: 38066362 Review.
-
Use of Bacteriophages to Control Vibrio Contamination of Microalgae Used as a Food Source for Oyster Larvae During Hatchery Culture.Curr Microbiol. 2020 Aug;77(8):1811-1820. doi: 10.1007/s00284-020-01981-w. Epub 2020 Apr 24. Curr Microbiol. 2020. PMID: 32328752
References
-
- Ackermann, H. W., and M. S. Dubow. 1987. Viruses of prokaryotes, vol. II. Natural groups of bacteriophages. CRC Press, Inc., Boca Raton, Fla.
-
- Barrow, P. A., and J. S. Soothill. 1997. Bacteriophage therapy and prophylaxis: rediscovery and renewed assessment of potential. Trends Microbiol. 5:268-271. - PubMed
-
- Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. - PubMed
-
- Chen, J., and M. W. Griffiths. 1996. Salmonella detection in eggs using lux+ bacteriophages. J. Food Prot. 59:908-914. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

