Quantitative assessment of picoeukaryotes in the natural environment by using taxon-specific oligonucleotide probes in association with tyramide signal amplification-fluorescence in situ hybridization and flow cytometry
- PMID: 12957941
- PMCID: PMC194996
- DOI: 10.1128/AEM.69.9.5519-5529.2003
Quantitative assessment of picoeukaryotes in the natural environment by using taxon-specific oligonucleotide probes in association with tyramide signal amplification-fluorescence in situ hybridization and flow cytometry
Abstract
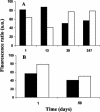
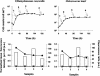
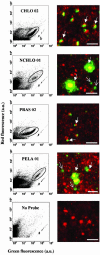
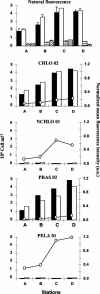
Picoeukaryotes (cells of <3 micro m in diameter) contribute significantly to marine plankton biomass and productivity, and recently molecular studies have brought to light their wide diversity. Among the methods that have been used so far to quantify aquatic microorganisms, fluorescence in situ hybridization of oligonucleotide probes combined with flow cytometry offers the advantages of both high resolution for taxonomic identification and automated cell counting. However, cell losses, cell clumps, and low signal-to-background ratio have often been mentioned as major problems for routine application of this combination of techniques. We developed a new protocol associating tyramide signal amplification-fluorescence in situ hybridization and flow cytometry, which allows the detection of picoeukaryotes in cultures during both the exponential and stationary phases. The use of surfactant and sonication proved to be essential for the detection and quantification of picoeukaryotes from the natural environment, with as little as a few tenths of a milliliter of 3- micro m-pore-size prefiltered sea water. The routine application of the technique was tested along a coastal transect off Brittany (France), where the different groups of picoeukaryotes were investigated using already published specific probes and a newly designed probe that targets the order Mamiellales (Prasinophyceae, Chlorophyta). Among the picoeukaryotes, Mamiellales outnumbered by 1 order of magnitude both the cyanobacteria and the non-Chlorophyta, which were represented mainly by the Pelagophyceae class. Picoeukaryote abundance increased from open toward more estuarine water, probably following changes in water temperature and stability.
Figures


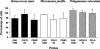
References
-
- Andersen, R. A., G. W. Saunders, M. P. Paskind, and J. P. Sexton. 1993. Ultrastructure and 18S rRNA gene sequence for Pelagomonas calceolata gen. et sp. nov. and the description of a new algal class, the Pelagophyceae classis nov. J. Phycol. 29:701-715.
-
- Båmstedt, U., and H. R. Skjoldal. 1976. Studies on the deep water pelagic community of Korfjorden, western Norway. Adenosine phosphate and nucleic acids in Euchaeta norvegica (Copepoda) in relation to its life cycle. Sarsia 60:63-80.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

