Plasmid transfer from Pseudomonas putida to the indigenous bacteria on alfalfa sprouts: characterization, direct quantification, and in situ location of transconjugant cells
- PMID: 12957943
- PMCID: PMC194921
- DOI: 10.1128/AEM.69.9.5536-5542.2003
Plasmid transfer from Pseudomonas putida to the indigenous bacteria on alfalfa sprouts: characterization, direct quantification, and in situ location of transconjugant cells
Abstract
The transfer of the plasmids pJKJ5 and TOL (pWWO) from Pseudomonas putida to the indigenous bacterial community on alfalfa sprouts was studied. Tagging with fluorescent protein markers allowed direct quantification of the introduced donor bacteria and of indigenous bacteria that had received the plasmids. The sprouts were observed for 9 days; during this time alfalfa seeds, inoculated with donor bacteria, developed to edible and subsequently decaying sprouts. The first transconjugants were detected on day 6 after donor inoculation and occurred at frequencies of 3.4 x 10(-4) and 2.0 x 10(-6) transconjugant cells per donor cell for pKJK5::gfp and TOL::gfp, respectively. Confocal laser scanning microscopy revealed that the sprouts were heavily colonized with donors and that most transconjugants were located around the hypocotyl and root areas. Randomly selected members of the indigenous bacterial community from both inoculated and uninoculated sprouts, as well as a representative part of the community that had received the plasmids, were characterized by polymorphisms of PCR-amplified ribosomal DNA (rDNA) spacer regions between the 16S and 23S genes, followed by partial 16S rDNA sequencing. This showed that the initially dominating genera Erwinia and Paenibacillus were gradually replaced by Pseudomonas on the fully developed sprouts. Transconjugants carrying either of the investigated plasmids mainly belonged to the genera Pseudomonas and ERWINIA: The numbers of transconjugant cells did not reach detectable levels until 6 days after the onset of germination, at which point these species constituted the majority of the indigenous bacteria. In conclusion, the alfalfa sprouts provided an environment that allowed noteworthy frequencies of plasmid transfer from P. putida in the absence of selective pressure that could favor the presence of the investigated plasmids.
Figures

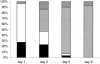
 ), Erwinia (□), and Paenibacillus (▪) spp. and for species either not identified or present in proportions of <5% (▨) are presented.
), Erwinia (□), and Paenibacillus (▪) spp. and for species either not identified or present in proportions of <5% (▨) are presented.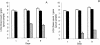
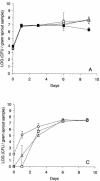
 ) counted in sprout samples at days 1, 6, and 9 after inoculation with donor cells containing either pKJK5::gfp (A) or TOL::gfp (B). The error bars represent the standard errors.
) counted in sprout samples at days 1, 6, and 9 after inoculation with donor cells containing either pKJK5::gfp (A) or TOL::gfp (B). The error bars represent the standard errors.
Similar articles
-
Establishment of new genetic traits in a microbial biofilm community.Appl Environ Microbiol. 1998 Jun;64(6):2247-55. doi: 10.1128/AEM.64.6.2247-2255.1998. Appl Environ Microbiol. 1998. PMID: 9603843 Free PMC article.
-
Dual labeling of Pseudomonas putida with fluorescent proteins for in situ monitoring of conjugal transfer of the TOL plasmid.Appl Environ Microbiol. 2003 Aug;69(8):4846-52. doi: 10.1128/AEM.69.8.4846-4852.2003. Appl Environ Microbiol. 2003. PMID: 12902279 Free PMC article.
-
Conjugational transfer of recombinant DNA in cultures and in soils: host range of Pseudomonas putida TOL plasmids.Appl Environ Microbiol. 1991 Oct;57(10):3020-7. doi: 10.1128/aem.57.10.3020-3027.1991. Appl Environ Microbiol. 1991. PMID: 1660698 Free PMC article.
-
Monitoring the conjugal transfer of plasmid RP4 in activated sludge and in situ identification of the transconjugants.FEMS Microbiol Lett. 1999 May 1;174(1):9-17. doi: 10.1111/j.1574-6968.1999.tb13543.x. FEMS Microbiol Lett. 1999. PMID: 10234817
-
Quantification of the presence and activity of specific microorganisms in nature.Mol Biotechnol. 1997 Apr;7(2):103-20. doi: 10.1007/BF02761746. Mol Biotechnol. 1997. PMID: 9219225 Review.
Cited by
-
Experimental approaches to tracking mobile genetic elements in microbial communities.FEMS Microbiol Rev. 2020 Sep 1;44(5):606-630. doi: 10.1093/femsre/fuaa025. FEMS Microbiol Rev. 2020. PMID: 32672812 Free PMC article. Review.
-
Novel assay to assess permissiveness of a soil microbial community toward receipt of mobile genetic elements.Appl Environ Microbiol. 2010 Jul;76(14):4813-8. doi: 10.1128/AEM.02713-09. Epub 2010 May 28. Appl Environ Microbiol. 2010. PMID: 20511430 Free PMC article.
-
Green fluorescent protein-labeled monitoring tool to quantify conjugative plasmid transfer between Gram-positive and Gram-negative bacteria.Appl Environ Microbiol. 2012 Feb;78(3):895-9. doi: 10.1128/AEM.05578-11. Epub 2011 Dec 2. Appl Environ Microbiol. 2012. PMID: 22138997 Free PMC article.
-
Characterization and Transferability of erm and tet Antibiotic Resistance Genes in Lactobacillus spp. Isolated from Traditional Fermented Milk.Curr Microbiol. 2022 Oct 8;79(11):339. doi: 10.1007/s00284-022-02980-9. Curr Microbiol. 2022. PMID: 36209320
-
Novel Butane-Oxidizing Bacteria and Diversity of bmoX Genes in Puguang Gas Field.Front Microbiol. 2018 Jul 17;9:1576. doi: 10.3389/fmicb.2018.01576. eCollection 2018. Front Microbiol. 2018. PMID: 30065710 Free PMC article.
References
-
- Bagdasarian, M., R. Lurz, B. Ruckert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. - PubMed
-
- Björklöf, K., E.-L. Nurmiaho-Lassila, N. Klinger, K. Haahtela, and M. Romantschuk. 2000. Colonization strategies and conjugal gene transfer of inoculated Pseudomonas syringae on the leaf surface. J. Appl. Microbiol. 89:423-432. - PubMed
-
- Bower, C. K., and M. A. Daeschel. 1999. Resistance responses of microorganisms in food environments. Int. J. Food Microbiol. 50:33-44. - PubMed
-
- Bradley, D. E. 1983. Specification of the conjugative pili and surface mating systems of Pseudomonas plasmids. J. Gen. Microbiol. 129:2545-2556. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

