Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea)
- PMID: 12957957
- PMCID: PMC194984
- DOI: 10.1128/AEM.69.9.5656-5663.2003
Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea)
Abstract
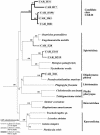
Present knowledge of microbial diversity is decidedly incomplete (S. J. Giovannoni and M. S. Rappé, p. 47-84, in D. Kirchman, ed., Microbial Ecology of the Oceans, 2000; E. Stackebrandt and T. M. Embley, p. 57-75, in R. R. Colwell and D. J. Grimes, ed., Nonculturable Microorganisms in the Environment, 2000). Protistan phylogenies are particularly deficient and undoubtedly exclude clades of principal ecological and evolutionary importance (S. L. Baldauf, Science 300:1703-1706, 2003). The rRNA approach has been extraordinarily successful in expanding the global prokaryotic record (S. J. Giovannoni and M. S. Rappé, p. 47-84, in D. Kirchman, ed., Microbial Ecology of the Oceans, 2000; E. Stackebrandt and T. M. Embley, p. 57-75, in R. R. Colwell and D. J. Grimes, ed., Nonculturable Microorganisms in the Environment, 2000) but has rarely been used in protistan discovery. Here we report the first application of the 18S rRNA approach to a permanently anoxic environment, the Cariaco Basin off the Venezuelan coast. On the basis of rRNA sequences, we uncovered a substantial number of novel protistan lineages. These included new clades of the highest taxonomic level unrelated to any known eukaryote as well as deep branches within established protistan groups. Three novel lineages branch at the base of the eukaryotic evolutionary tree preceding, contemporary with, or immediately following the earliest eukaryotic branches. These newly discovered protists may retain traits reminiscent of an early eukaryotic ancestor(s).
Figures




Similar articles
-
A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples.Protist. 2006 Feb;157(1):31-43. doi: 10.1016/j.protis.2005.10.004. Epub 2006 Jan 23. Protist. 2006. PMID: 16431157
-
Protistan microbial observatory in the Cariaco Basin, Caribbean. I. Pyrosequencing vs Sanger insights into species richness.ISME J. 2011 Aug;5(8):1344-56. doi: 10.1038/ismej.2011.6. Epub 2011 Mar 10. ISME J. 2011. PMID: 21390079 Free PMC article.
-
Hydroids (Cnidaria, Hydrozoa) from Mauritanian Coral Mounds.Zootaxa. 2020 Nov 16;4878(3):zootaxa.4878.3.2. doi: 10.11646/zootaxa.4878.3.2. Zootaxa. 2020. PMID: 33311142
-
Evolution of the protists and protistan parasites from the perspective of molecular systematics.Int J Parasitol. 1998 Jan;28(1):11-20. doi: 10.1016/s0020-7519(97)00181-1. Int J Parasitol. 1998. PMID: 9504331 Review.
-
Unveiling new microbial eukaryotes in the surface ocean.Curr Opin Microbiol. 2008 Jun;11(3):213-8. doi: 10.1016/j.mib.2008.04.004. Epub 2008 Jun 13. Curr Opin Microbiol. 2008. PMID: 18556239 Review.
Cited by
-
Diversity of microbial eukaryotes in sediment at a deep-sea methane cold seep: surveys of ribosomal DNA libraries from raw sediment samples and two enrichment cultures.Extremophiles. 2007 Jul;11(4):563-76. doi: 10.1007/s00792-007-0068-z. Epub 2007 Apr 11. Extremophiles. 2007. PMID: 17426921
-
Protistan community patterns within the brine and halocline of deep hypersaline anoxic basins in the eastern Mediterranean Sea.Extremophiles. 2009 Jan;13(1):151-67. doi: 10.1007/s00792-008-0206-2. Epub 2008 Dec 5. Extremophiles. 2009. PMID: 19057844
-
Eukaryotic diversity in an anaerobic aquifer polluted with landfill leachate.Appl Environ Microbiol. 2008 Jul;74(13):3959-68. doi: 10.1128/AEM.02820-07. Epub 2008 May 9. Appl Environ Microbiol. 2008. PMID: 18469120 Free PMC article.
-
Microeukaryotic community and oxygen response in rice field soil revealed using a combined rRNA-gene and rRNA-based approach.Microbes Environ. 2014;29(1):74-81. doi: 10.1264/jsme2.me13128. Epub 2014 Feb 7. Microbes Environ. 2014. PMID: 24521691 Free PMC article.
-
Evidence from the resurrected family Polyrhabdinidae Kamm, 1922 (Apicomplexa: Gregarinomorpha) supports the epimerite, an attachment organelle, as a major eugregarine innovation.PeerJ. 2021 Sep 16;9:e11912. doi: 10.7717/peerj.11912. eCollection 2021. PeerJ. 2021. PMID: 34616591 Free PMC article.
References
-
- Amaral Zettler, L. A., F. Goméz, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Microbiology: eukaryotic diversity in Spain's River of Fire. Nature 417:137. - PubMed
-
- Baldauf, S. L. 2003. The deep roots of eukaryotes. Science 300:1703-1706. - PubMed
-
- Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Müller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

