Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory (an 18-month evaluation)
- PMID: 12958237
- PMCID: PMC193817
- DOI: 10.1128/JCM.41.9.4134-4140.2003
Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory (an 18-month evaluation)
Abstract
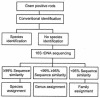
We have evaluated over a period of 18 months the use of 16S ribosomal DNA (rDNA) sequence analysis as a means of identifying aerobic gram-positive rods in the clinical laboratory. Two collections of strains were studied: (i) 37 clinical strains of gram-positive rods well identified by phenotypic tests, and (ii) 136 clinical isolates difficult to identify by standard microbiological investigations, i.e., identification at the species level was impossible. Results of molecular analyses were compared with those of conventional phenotypic identification procedures. Good overall agreement between phenotypic and molecular identification procedures was found for the collection of 37 clinical strains well identified by conventional means. For the 136 clinical strains which were difficult to identify by standard microbiological investigations, phenotypic characterization identified 71 of 136 (52.2%) isolates at the genus level; 65 of 136 (47.8%) isolates could not be discriminated at any taxonomic level. In comparison, 16S rDNA sequencing identified 89 of 136 (65.4%) isolates at the species level, 43 of 136 (31.6%) isolates at the genus level, and 4 of 136 (2.9%) isolates at the family level. We conclude that (i) rDNA sequencing is an effective means for the identification of aerobic gram-positive rods which are difficult to identify by conventional techniques, and (ii) molecular identification procedures are not required for isolates well identified by phenotypic investigations.
Figures
References
-
- Boettger, E. C. 1989. Rapid determination of bacterial ribosomal RNA sequences by direct sequencing of enzymatically amplified DNA. FEMS Microbiol. Lett. 65:171-176. - PubMed
-
- Boettger, E. C. 1996. Approaches for identification of microorganisms. ASM News 62:247-250.
-
- Bosshard, P. P., R. Zbinden, and M. Alwegg. 2002. Paenibacillus turicensis sp. nov., a novel bacterium harboring heterogeneities between 16S rRNA genes. Int. J. Syst. E vol. Microbiol. 52:2241-2249. - PubMed
-
- Clayton, R. A., G. Sutton, P. S. Hinkle, C. Bult, and C. Fields. 1995. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol. 45:595-599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources