Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis
- PMID: 12958269
- PMCID: PMC193777
- DOI: 10.1128/JCM.41.9.4359-4365.2003
Use of loop-mediated isothermal amplification of the IS900 sequence for rapid detection of cultured Mycobacterium avium subsp. paratuberculosis
Abstract
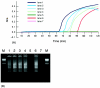
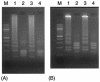
We evaluated the usefulness of loop-mediated isothermal amplification (LAMP) in detecting specific gene sequences of Mycobacterium avium subsp. paratuberculosis (MAP). A total of 102 primer sets for LAMP was designed to amplify the IS900, HspX, and F57 gene sequences of MAP. Using each of two primer sets (P-1 and P-2) derived from the IS900 fragment, it was possible to detect MAP in a manner similar to that used with nested PCR. The sensitivity of LAMP with P-1 was 0.5 pg/tube, which was more sensitive than nested PCR. When P-2 was used, 5 pg/tube could be detected, which was the same level of sensitivity as that for nested PCR. LAMP with P-1 was specific. Although only 2 Mycobacterium scrofulaceum strains out of 43 non-MAP mycobacterial strains were amplified, the amplification reaction for these strains was less efficient than for MAP strains, and their products could be distinguished from MAP products by restriction digestion. LAMP with P-2 resulted in very specific amplification only from MAP, the same result obtained with nested PCR. Our LAMP method was highly specific, and the white turbidity of magnesium pyrophosphate, a by-product of the LAMP reaction, allowed simple visual detection. Our method is rapid, taking only 2 h, compared with 4 h for nested PCR. In addition, the LAMP method is performed under isothermal conditions and no special apparatus is needed, which makes it more economical and practical than nested PCR or real-time PCR. These results indicate that LAMP can provide a rapid yet simple test for the detection of MAP.
Figures




References
-
- Bannantine, J. P., and J. R. Stable. 2000. HspX is present within Mycobacterium paratubercurosis-infected macrophages and is recognized by sera from some infected cattle. Vet. Microbiol. 76:343-358. - PubMed
-
- Beard, P. M., D. Henderson, M. J. Daniels, A. Pirie, D. Buxton, A. Greig, M. R. Hutchings, I. McHendrick, S. Rhind, K. Stevenson, and J. M. Sharp. 1999. Evidence of paratuberculosis in fox (Vulpes vulpes) and stoat (Mustelaerminea). Vet. Rec. 145:612-613.
-
- Chiodini, R. J., H. J. Van Kruiningen, and R. S. Merkal. 1984. Ruminant paratuberculosis (Johne's disease): the current status and future prospect. Cornell Vet. 74:218-262. - PubMed
-
- Collins, D. M., D. M. Stephens, and G. W. De Lisle. 1993. Comparison of polymerase chain reaction tests and fecal culture for detecting Mycobacterium paratuberculosis in bovine feces. Vet. Microbiol. 36:289-299. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

