Strand-biased DNA methylation associated with centromeric regions in Arabidopsis
- PMID: 12960391
- PMCID: PMC196939
- DOI: 10.1073/pnas.1831011100
Strand-biased DNA methylation associated with centromeric regions in Arabidopsis
Abstract
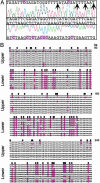
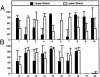
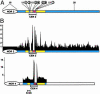
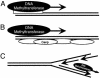
The Arabidopsis genome project assembled 15 megabases of heterochromatic sequence, facilitating investigations of heterochromatin assembly, maintenance, and structure. In many species, large quantities of methylcytosine decorate heterochromatin; these modifications are typically maintained by methyltransferases that recognize newly replicated hemimethylated DNA. We assessed the extent and patterns of Arabidopsis heterochromatin methylation by amplifying and sequencing genomic DNA treated with bisulfite, which converts cytosine, but not methylcytosine, to uracil. This survey revealed unexpected asymmetries in methylation patterns, with one helix strand often exhibiting higher levels of methylation. We confirmed these observations both by immunoprecipitating methylated DNA strands and by restriction enzyme digestion of amplified, bisulfite-treated DNA. We also developed a primer-extension assay that can monitor the methylation status of an entire chromosome, demonstrating that strand-specific methylation occurs predominantly in the centromeric regions. Conventional models for methylation maintenance do not explain these unusual patterns; instead, new models that allow for strand specificity are required. The abundance of Arabidopsis strand-specific modifications points to their importance, perhaps as epigenetic signals that mark the heterochromatic regions that confer centromere activity.
Figures


Similar articles
-
Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1.Science. 2002 Sep 13;297(5588):1871-3. doi: 10.1126/science.1074950. Epub 2002 Jun 20. Science. 2002. PMID: 12077425
-
Bisulphite sequencing of plant genomic DNA.Methods Mol Biol. 2010;655:433-43. doi: 10.1007/978-1-60761-765-5_29. Methods Mol Biol. 2010. PMID: 20734278
-
Distribution of 5-methylcytosine residues in 5S rRNA genes in Arabidopsis thaliana and Secale cereale.Mol Genet Genomics. 2002 Dec;268(4):510-7. doi: 10.1007/s00438-002-0761-7. Epub 2002 Oct 25. Mol Genet Genomics. 2002. PMID: 12471448
-
Methylation and demethylation of the Arabidopsis genome.Curr Opin Plant Biol. 2011 Apr;14(2):137-41. doi: 10.1016/j.pbi.2010.11.004. Epub 2010 Dec 13. Curr Opin Plant Biol. 2011. PMID: 21159546 Review.
-
DNA methylation dynamics in plant genomes.Biochim Biophys Acta. 2007 May-Jun;1769(5-6):276-86. doi: 10.1016/j.bbaexp.2007.01.009. Epub 2007 Feb 7. Biochim Biophys Acta. 2007. PMID: 17341434 Review.
Cited by
-
Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation.PLoS Genet. 2006 Feb;2(2):e17. doi: 10.1371/journal.pgen.0020017. Epub 2006 Feb 10. PLoS Genet. 2006. PMID: 16477312 Free PMC article.
-
Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats.PLoS Genet. 2005 Dec;1(6):e79. doi: 10.1371/journal.pgen.0010079. Epub 2005 Dec 23. PLoS Genet. 2005. PMID: 16389298 Free PMC article.
-
Genome-wide mapping of DNA methylation in the human malaria parasite Plasmodium falciparum.Cell Host Microbe. 2013 Dec 11;14(6):696-706. doi: 10.1016/j.chom.2013.11.007. Cell Host Microbe. 2013. PMID: 24331467 Free PMC article.
-
Cytological investigation of Haplopappus gracilis (Nutt.) Gray: 5-methylcytosine-rich regions, fluorochrome banding and chromatin sensitivity to DNase I digestion.Protoplasma. 2008;233(1-2):107-13. doi: 10.1007/s00709-008-0296-9. Epub 2008 Jul 10. Protoplasma. 2008. PMID: 18615238
-
Changes in global gene expression in response to chemical and genetic perturbation of chromatin structure.PLoS One. 2011;6(6):e20587. doi: 10.1371/journal.pone.0020587. Epub 2011 Jun 3. PLoS One. 2011. PMID: 21673996 Free PMC article.
References
-
- Jones, P. L. & Wolffe, A. P. (1999) Semin. Cancer Biol. 9, 339-347. - PubMed
-
- Gruenbaum, Y., Stein, R., Cedar, H. & Razin, A. (1981) FEBS Lett. 124, 67-71. - PubMed
-
- Ng, H.-H. & Bird, A. P. (1999) Curr. Opin. Genet. Dev. 9, 158-163. - PubMed
-
- Robertson, K. D. & Jones, P. A. (2000) Carcinogenesis 21, 461-467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

