Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration
- PMID: 12960403
- PMCID: PMC196873
- DOI: 10.1073/pnas.1834348100
Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration
Abstract
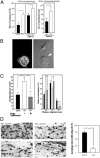
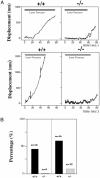
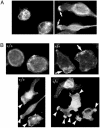
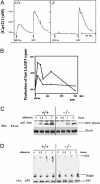
The biological role of the protein tyrosine kinase, Pyk2, was explored by targeting the Pyk2 gene by homologous recombination. Pyk2-/- mice are viable and fertile, without overt impairment in development or behavior. However, the morphology and behavior of Pyk2-/- macrophages were impaired. Macrophages isolated from mutant mice failed to become polarized, to undergo membrane ruffling, and to migrate in response to chemokine stimulation. Moreover, the contractile activity in the lamellipodia of Pyk2-/- macrophages was impaired, as revealed by measuring the rearward movement toward the nucleus of fibronectin-coated beads on the lamellipodia in opposition to an immobilizing force generated by optical tweezers. Consistently, the infiltration of macrophages into a carageenan-induced inflammatory region was strongly inhibited in Pyk2-/- mice. In addition, chemokine stimulation of inositol (1, 4, 5) triphosphate production and Ca2+ release, as well as integrin-induced activation of Rho and phosphatidyl inositol 3 kinase, were compromised in Pyk2-/- macrophages. These experiments reveal a role for Pyk2 in cell signaling in macrophages essential for cell migration and function.
Figures

References
-
- Lev, S., Moreno, H., Martinez, R., Canoll, P., Peles, E., Musacchio, J. M., Plowman, G. D., Rudy, B. & Schlessinger, J. (1995) Nature 376, 737–745. - PubMed
-
- Avraham, S., London, R., Fu, Y., Ota, S., Hiregowdara, D., Li, J., Jiang, S., Pasztor, L. M., White, R. A., Groopman, J. E. & Avraham, H. (1995) J. Biol. Chem. 270, 27742–27751. - PubMed
-
- Sasaki, H., Nagura, K., Ishino, M., Tobioka, H., Kotani, K. & Sasaki, T. (1995) J. Biol. Chem. 270, 21206–21219. - PubMed
-
- Yu, H., Li, X., Marchetto, G. S., Dy, R., Hunter, D., Calvo, B., Dawson, T. L., Wilm, M., Anderegg, R. J., Graves, L. M. & Earp, H. S. (1996) J. Biol. Chem. 271, 29993–29998. - PubMed
-
- Girault, J. A., Costa, A., Derkinderen, P., Studler, J. M. & Toutant, M. (1999) Trends Neurosci. 22, 257–263. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

