Coarse-grained sequences for protein folding and design
- PMID: 12963815
- PMCID: PMC196869
- DOI: 10.1073/pnas.1931882100
Coarse-grained sequences for protein folding and design
Abstract
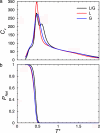
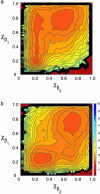
We present the results of sequence design on our off-lattice minimalist model in which no specification of native-state tertiary contacts is needed. We start with a sequence that adopts a target topology and build on it through sequence mutation to produce new sequences that comprise distinct members within a target fold class. In this work, we use the alpha/beta ubiquitin fold class and design two new sequences that, when characterized through folding simulations, reproduce the differences in folding mechanism seen experimentally for proteins L and G. The primary implication of this work is that patterning of hydrophobic and hydrophilic residues is the physical origin for the success of relative contact-order descriptions of folding, and that these physics-based potentials provide a predictive connection between free energy landscapes and amino acid sequence (the original protein folding problem). We present results of the sequence mapping from a 20- to the three-letter code for determining a sequence that folds into the WW domain topology to illustrate future extensions to protein design.
Figures



References
-
- Onuchic, J. N., Lutheyschulten, Z. & Wolynes, P. G. (1997) Annu. Rev. Phys. Chem. 48, 545–600. - PubMed
-
- Go, N. (1983) Annu. Rev. Biophys. Bioeng. 12, 183–210. - PubMed
-
- Koga, N. & Takada, S. (2001) J. Mol. Biol. 313, 171–180. - PubMed
-
- Plaxco, K. W., Simons, K. T., Ruczinski, I. & David, B. (2000) Biochemistry 39, 11177–11183. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

