Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187
- PMID: 12963818
- PMCID: PMC196855
- DOI: 10.1073/pnas.1832879100
Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187
Erratum in
- Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):14510
Abstract
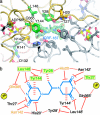
Type IIA topoisomerases both manage the topological state of chromosomal DNA and are the targets of a variety of clinical agents. Bisdioxopiperazines are anticancer agents that associate with ATP-bound eukaryotic topoisomerase II (topo II) and convert the enzyme into an inactive, salt-stable clamp around DNA. To better understand both topo II and bisdioxopiperazine function, we determined the structures of the adenosine 5'-[beta,gamma-imino]-triphosphate-bound yeast topo II ATPase region (ScT2-ATPase) alone and complexed with the bisdioxopiperazine ICRF-187. The drug-free form of the protein is similar in overall fold to the equivalent region of bacterial gyrase but unexpectedly displays significant conformational differences. The ternary drug-bound complex reveals that ICRF-187 acts by an unusual mechanism of inhibition in which the drug does not compete for the ATP-binding pocket, but bridges and stabilizes a transient dimer interface between two ATPase protomers. Our data explain why bisdioxopiperazines target ATP-bound topo II, provide a structural rationale for the effects of certain drug-resistance mutations, and point to regions of bisdioxopiperazines that might be modified to improve or alter drug specificity.
Figures



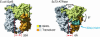
Similar articles
-
Human small cell lung cancer NYH cells selected for resistance to the bisdioxopiperazine topoisomerase II catalytic inhibitor ICRF-187 demonstrate a functional R162Q mutation in the Walker A consensus ATP binding domain of the alpha isoform.Cancer Res. 1999 Jul 15;59(14):3442-50. Cancer Res. 1999. PMID: 10416608
-
Chinese hamster ovary cells resistant to the topoisomerase II catalytic inhibitor ICRF-159: a Tyr49Phe mutation confers high-level resistance to bisdioxopiperazines.Cancer Res. 1998 Apr 1;58(7):1460-8. Cancer Res. 1998. PMID: 9537249
-
Probing the interaction of the cytotoxic bisdioxopiperazine ICRF-193 with the closed enzyme clamp of human topoisomerase IIalpha.Mol Pharmacol. 2000 Sep;58(3):560-8. doi: 10.1124/mol.58.3.560. Mol Pharmacol. 2000. PMID: 10953049
-
Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism.Biochem Soc Trans. 2005 Dec;33(Pt 6):1465-70. doi: 10.1042/BST0331465. Biochem Soc Trans. 2005. PMID: 16246147 Review.
-
DNA supercoiling and relaxation by ATP-dependent DNA topoisomerases.Philos Trans R Soc Lond B Biol Sci. 1992 Apr 29;336(1276):83-91. doi: 10.1098/rstb.1992.0047. Philos Trans R Soc Lond B Biol Sci. 1992. PMID: 1351300 Review.
Cited by
-
Evolutionary History of TOPIIA Topoisomerases in Animals.J Mol Evol. 2022 Apr;90(2):149-165. doi: 10.1007/s00239-022-10048-2. Epub 2022 Feb 14. J Mol Evol. 2022. PMID: 35165762
-
Anthracyclines as Topoisomerase II Poisons: From Early Studies to New Perspectives.Int J Mol Sci. 2018 Nov 6;19(11):3480. doi: 10.3390/ijms19113480. Int J Mol Sci. 2018. PMID: 30404148 Free PMC article. Review.
-
Structural insights into fungal and human topoisomerase II with implications for in silico antifungal drug design.Sci Rep. 2025 Mar 19;15(1):9467. doi: 10.1038/s41598-025-93122-1. Sci Rep. 2025. PMID: 40108235 Free PMC article.
-
Targeting DNA Topoisomerase II in Antifungal Chemotherapy.Molecules. 2022 Nov 11;27(22):7768. doi: 10.3390/molecules27227768. Molecules. 2022. PMID: 36431868 Free PMC article. Review.
-
Solution structures of DNA-bound gyrase.Nucleic Acids Res. 2011 Jan;39(2):755-66. doi: 10.1093/nar/gkq799. Epub 2010 Sep 24. Nucleic Acids Res. 2011. PMID: 20870749 Free PMC article.
References
-
- Wang, J. C. (1998) Q. Rev. Biophys. 31, 107–144. - PubMed
-
- Lynn, R., Giaever, G., Swanberg, S. L. & Wang, J. C. (1986) Science 233, 647–649. - PubMed
-
- Peng, H. & Marians, K. J. (1993) J. Biol. Chem. 268, 24481–24490. - PubMed
-
- Berger, J. M., Gamblin, S. J., Harrison, S. C. & Wang, J. C. (1996) Nature 379, 225–232. - PubMed
-
- Osheroff, N. (1986) J. Biol. Chem. 261, 9944–9950. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

