Ancient origin of the tryptophan operon and the dynamics of evolutionary change
- PMID: 12966138
- PMCID: PMC193870
- DOI: 10.1128/MMBR.67.3.303-342.2003
Ancient origin of the tryptophan operon and the dynamics of evolutionary change
Abstract
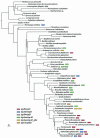
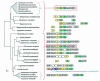
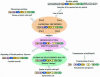
The seven conserved enzymatic domains required for tryptophan (Trp) biosynthesis are encoded in seven genetic regions that are organized differently (whole-pathway operons, multiple partial-pathway operons, and dispersed genes) in prokaryotes. A comparative bioinformatics evaluation of the conservation and organization of the genes of Trp biosynthesis in prokaryotic operons should serve as an excellent model for assessing the feasibility of predicting the evolutionary histories of genes and operons associated with other biochemical pathways. These comparisons should provide a better understanding of possible explanations for differences in operon organization in different organisms at a genomics level. These analyses may also permit identification of some of the prevailing forces that dictated specific gene rearrangements during the course of evolution. Operons concerned with Trp biosynthesis in prokaryotes have been in a dynamic state of flux. Analysis of closely related organisms among the Bacteria at various phylogenetic nodes reveals many examples of operon scission, gene dispersal, gene fusion, gene scrambling, and gene loss from which the direction of evolutionary events can be deduced. Two milestone evolutionary events have been mapped to the 16S rRNA tree of Bacteria, one splitting the operon in two, and the other rejoining it by gene fusion. The Archaea, though less resolved due to a lesser genome representation, appear to exhibit more gene scrambling than the Bacteria. The trp operon appears to have been an ancient innovation; it was already present in the common ancestor of Bacteria and Archaea. Although the operon has been subjected, even in recent times, to dynamic changes in gene rearrangement, the ancestral gene order can be deduced with confidence. The evolutionary history of the genes of the pathway is discernible in rough outline as a vertical line of descent, with events of lateral gene transfer or paralogy enriching the analysis as interesting features that can be distinguished. As additional genomes are thoroughly analyzed, an increasingly refined resolution of the sequential evolutionary steps is clearly possible. These comparisons suggest that present-day trp operons that possess finely tuned regulatory features are under strong positive selection and are able to resist the disruptive evolutionary events that may be experienced by simpler, poorly regulated operons.
Figures













References
-
- Ahmad, S., and R. A. Jensen. 1989. Utility of a bifunctional tryptophan pathway enzyme for the classification of the Herbicola-Agglomerans complex of bacteria. Int. J. Syst. Bacteriol. 39:100-104.
-
- Altamirano, M. M., J. M. Blackburn, C. Aguayo, and A. R. Fersht. 2000. Directed evolution of new catalytic activity with the α/β-barrel scaffold. Nature 403:617-622. - PubMed
-
- Altamirano, N., J. M. Blackburn, C. Aguayo, and A. R. Fersht. 2002. Directed evolution of new catalytic activity with the α/β-barrel scaffold: retraction. Nature 417:468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

