Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
- PMID: 12970175
- PMCID: PMC212723
- DOI: 10.1093/emboj/cdg456
Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Abstract
The nuclear receptor heterodimers of liver X receptor (LXR) and retinoid X receptor (RXR) are key transcriptional regulators of genes involved in lipid homeostasis and inflammation. We report the crystal structure of the ligand-binding domains (LBDs) of LXRalpha and RXRbeta complexed to the synthetic LXR agonist T-0901317 and the RXR agonist methoprene acid (Protein Data Base entry 1UHL). Both LBDs are in agonist conformation with GRIP-1 peptides bound at the coactivator binding sites. T-0901317 occupies the center of the LXR ligand-binding pocket and its hydroxyl head group interacts with H421 and W443, residues identified by mutational analysis as critical for ligand-induced transcriptional activation by T-0901317 and various endogenous oxysterols. The topography of the pocket suggests a common anchoring of these oxysterols via their 22-, 24- or 27-hydroxyl group to H421 and W443. Polyunsaturated fatty acids act as LXR antagonists and an E267A mutation was found to enhance their transcriptional inhibition. The present structure provides a powerful tool for the design of novel modulators that can be used to characterize further the physiological functions of the LXR-RXR heterodimer.
Figures

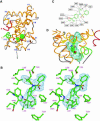




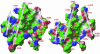
References
-
- Abagyan R.A., Totrov,M.M. and Kuznetsov,D.N. (1994) ICM—a new method for protein modeling and design. Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem., 15, 488–506.
-
- Anderson L.M., Choe,S.E., Yukhananov,R.Y., Hopfner,R.L., Church,G.M., Pratt,R.E. and Dzau,V.J. (2003) Identification of a novel set of genes regulated by a unique liver X receptor-α-mediated transcription mechanism. J. Biol. Chem., 278, 15252–15260. - PubMed
-
- Bourguet W., Ruff,M., Chambon,P., Gronemeyer,H. and Moras,D. (1995) Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature, 375, 377–382. - PubMed
-
- Bricogne G. (1993) Direct phase determination by entropy maximization and likelihood ranking: status report and perspectives. Acta Crystallogr. D, 49, 37–60. - PubMed
-
- Brzozowski A.M. et al. (1997) Molecular basis of agonism and antagonism in the oestrogen receptor. Nature, 389, 753–758. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

