Antigenic subclasses of polytropic murine leukemia virus (MLV) isolates reflect three distinct groups of endogenous polytropic MLV-related sequences in NFS/N mice
- PMID: 12970417
- PMCID: PMC228382
- DOI: 10.1128/jvi.77.19.10327-10338.2003
Antigenic subclasses of polytropic murine leukemia virus (MLV) isolates reflect three distinct groups of endogenous polytropic MLV-related sequences in NFS/N mice
Abstract
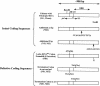
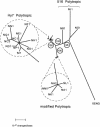
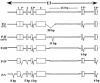
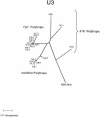
Polytropic murine leukemia viruses (MLVs) are generated by recombination of ecotropic MLVs with members of a family of endogenous proviruses in mice. Previous studies have indicated that polytropic MLV isolates comprise two mutually exclusive antigenic subclasses, each of which is reactive with one of two monoclonal antibodies termed MAb 516 and Hy 7. A major determinant of the epitopes distinguishing the subclasses mapped to a single amino acid difference in the SU protein. Furthermore, distinctly different populations of the polytropic MLV subclasses are generated upon inoculation of different ecotropic MLVs. Here we have characterized the majority of endogenous polytropic MLV-related proviruses of NFS/N mice. Most of the proviruses contain intact sequences encoding the receptor-binding region of the SU protein and could be distinguished by sequence heterogeneity within that region. We found that the endogenous proviruses comprise two major groups that encode the major determinant for Hy 7 or MAb 516 reactivity. The Hy 7-reactive proviruses correspond to previously identified polytropic proviruses, while the 516-reactive proviruses comprise the modified polytropic proviruses as well as a third group of polytropic MLV-related proviruses that exhibit distinct structural features. Phylogenetic analyses indicate that the latter proviruses reflect features of phylogenetic intermediates linking xenotropic MLVs to the polytropic and modified polytropic proviruses. These studies elucidate the relationships of the antigenic subclasses of polytropic MLVs to their endogenous counterparts, identify a new group of endogenous proviruses, and identify distinguishing characteristics of the proviruses that should facilitate a more precise description of their expression in mice and their participation in recombination to generate recombinant viruses.
Figures





Similar articles
-
Precise identification of endogenous proviruses of NFS/N mice participating in recombination with moloney ecotropic murine leukemia virus (MuLV) to generate polytropic MuLVs.J Virol. 2005 Apr;79(8):4664-71. doi: 10.1128/JVI.79.8.4664-4671.2005. J Virol. 2005. PMID: 15795252 Free PMC article.
-
Characterization of epitopes defining two major subclasses of polytropic murine leukemia viruses (MuLVs) which are differentially expressed in mice infected with different ecotropic MuLVs.J Virol. 1994 Aug;68(8):5194-203. doi: 10.1128/JVI.68.8.5194-5203.1994. J Virol. 1994. PMID: 7518532 Free PMC article.
-
Recombinant Origins of Pathogenic and Nonpathogenic Mouse Gammaretroviruses with Polytropic Host Range.J Virol. 2017 Oct 13;91(21):e00855-17. doi: 10.1128/JVI.00855-17. Print 2017 Nov 1. J Virol. 2017. PMID: 28794032 Free PMC article.
-
Genetics of endogenous murine leukemia viruses.Ann N Y Acad Sci. 1989;567:39-49. doi: 10.1111/j.1749-6632.1989.tb16457.x. Ann N Y Acad Sci. 1989. PMID: 2552892 Review.
-
The mouse "xenotropic" gammaretroviruses and their XPR1 receptor.Retrovirology. 2010 Nov 30;7:101. doi: 10.1186/1742-4690-7-101. Retrovirology. 2010. PMID: 21118532 Free PMC article. Review.
Cited by
-
Capsid-Targeted Viral Inactivation: A Novel Tactic for Inhibiting Replication in Viral Infections.Viruses. 2016 Sep 21;8(9):258. doi: 10.3390/v8090258. Viruses. 2016. PMID: 27657114 Free PMC article. Review.
-
Endogenous retroviruses mobilized during friend murine leukemia virus infection.Virology. 2016 Dec;499:136-143. doi: 10.1016/j.virol.2016.07.009. Epub 2016 Sep 19. Virology. 2016. PMID: 27657834 Free PMC article.
-
Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development.BMC Bioinformatics. 2010 Oct 25;11:530. doi: 10.1186/1471-2105-11-530. BMC Bioinformatics. 2010. PMID: 20973947 Free PMC article.
-
Precise identification of endogenous proviruses of NFS/N mice participating in recombination with moloney ecotropic murine leukemia virus (MuLV) to generate polytropic MuLVs.J Virol. 2005 Apr;79(8):4664-71. doi: 10.1128/JVI.79.8.4664-4671.2005. J Virol. 2005. PMID: 15795252 Free PMC article.
-
Origins of the endogenous and infectious laboratory mouse gammaretroviruses.Viruses. 2014 Dec 26;7(1):1-26. doi: 10.3390/v7010001. Viruses. 2014. PMID: 25549291 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

