An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector
- PMID: 12970424
- PMCID: PMC228391
- DOI: 10.1128/jvi.77.19.10394-10403.2003
An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector
Abstract
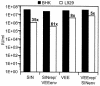
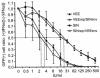
Alphavirus replicon particle-based vaccine vectors derived from Sindbis virus (SIN), Semliki Forest virus, and Venezuelan equine encephalitis virus (VEE) have been shown to induce robust antigen-specific cellular, humoral, and mucosal immune responses in many animal models of infectious disease and cancer. However, since little is known about the relative potencies among these different vectors, we compared the immunogenicity of replicon particle vectors derived from two very different parental alphaviruses, VEE and SIN, expressing a human immunodeficiency virus type 1 p55(Gag) antigen. Moreover, to explore the potential benefits of combining elements from different alphaviruses, we generated replicon particle chimeras of SIN and VEE. Two distinct strategies were used to produce particles with VEE-p55(gag) replicon RNA packaged within SIN envelope glycoproteins and SIN-p55(gag) replicon RNA within VEE envelope glycoproteins. Each replicon particle configuration induced Gag-specific CD8(+) T-cell responses in murine models when administered alone or after priming with DNA. However, Gag-specific responses varied dramatically, with the strongest responses to this particular antigen correlating with the VEE replicon RNA, irrespective of the source of envelope glycoproteins. Comparing the replicons with respect to heterologous gene expression levels and sensitivity to alpha/beta interferon in cultured cells indicated that each might contribute to potency differences. This work shows that combining desirable elements from VEE and SIN into a replicon particle chimera may be a valuable approach toward the goal of developing vaccine vectors with optimal in vivo potency, ease of production, and safety.
Figures





References
-
- Chastain, M., A. J. Simon, K. A. Soper, D. J. Holder, D. L. Montgomery, S. L. Sagar, D. R. Casimiro, and C. R. Middaugh. 2001. Antigen levels and antibody titers after DNA vaccination. J. Pharm. Sci. 90:474-484. - PubMed
-
- Choi, H. K., L. Tong, W. Minor, P. Dumas, U. Boege, M. G. Rossmann, and G. Wengler. 1991. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature 354:37-43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

