Transcription program of murine gammaherpesvirus 68
- PMID: 12970434
- PMCID: PMC228380
- DOI: 10.1128/jvi.77.19.10488-10503.2003
Transcription program of murine gammaherpesvirus 68
Abstract
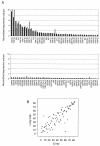
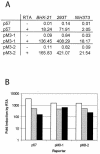
Murine gammaherpesvirus 68 (MHV-68 [also referred to as gammaHV68]) is phylogenetically related to Kaposi's sarcoma-associated herpesvirus (KSHV [also referred to as HHV-8]) and Epstein-Barr virus (EBV). However, unlike KSHV or EBV, MHV-68 readily infects fibroblast and epithelial cell lines derived from several mammalian species, providing a system to study productive and latent infections as well as reactivation of gammaherpesviruses in vivo and in vitro. To carry out rapid genome-wide analysis of MHV-68 gene expression, we made DNA arrays containing nearly all of the known and predicted open reading frames (ORFs) of the virus. RNA obtained from an MHV-68 latently infected cell line, from cells lytically infected with MHV-68 in culture, and from the lung tissue of infected mice was used to probe the MHV-68 arrays. Using a tightly latent B-cell line (S11E), the MHV-68 latent transcription program was quantitatively described. Using BHK-21 cells and infected mice, we demonstrated that latent genes are transcribed during lytic replication and are relatively independent of de novo protein synthesis. We determined that the transcription profiles at the peak of lytic gene expression are similar in cultured fibroblast and in the lung of infected mice. Finally, the MHV-68 DNA arrays were used to examine the gene expression profile of a recombinant virus that overexpresses replication and transcription activator (RTA), C-RTA/MHV-68, during lytic replication in cell culture. The recombinant virus replicates faster then the parental strain and the DNA arrays revealed that nearly every MHV-68 ORF examined was activated by RTA overexpression. Examination of the gene expression patterns of C-RTA/MHV-68 over a time course led to the finding that the M3 promoter is RTA responsive in the absence of other viral factors.
Figures

References
-
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

