Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase
- PMID: 12970436
- PMCID: PMC228489
- DOI: 10.1128/jvi.77.19.10515-10527.2003
Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase
Abstract
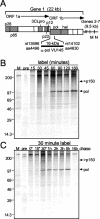
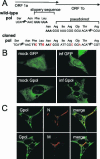
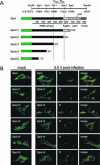
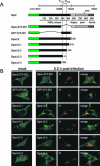
Mouse hepatitis virus (MHV) RNA synthesis is mediated by a viral RNA-dependent RNA polymerase (RdRp) on membrane-bound replication complexes in the host cell cytoplasm. However, it is not known how the putative MHV RdRp (Pol) is targeted to and retained on cellular membranes. In this report, we show that a 100-kDa protein was stably detected by an anti-Pol antiserum as a mature product throughout the virus life cycle. Gradient fractionation and biochemical extraction experiments demonstrated that Pol was not an integral membrane protein but was tightly associated with membranes and coimmunoprecipitated with the replicase proteins 3CLpro, p22, and p12. By immunofluorescence confocal microscopy, Pol colocalized with viral proteins at replication complexes, distinct from sites of virion assembly, over the entire course of infection. To determine if Pol associated with cellular membranes in the absence of other viral factors, the pol domain of gene 1 was cloned and expressed in cells as a fusion with green fluorescent protein, termed Gpol. In Gpol-expressing cells that were infected with MHV, but not in mock-infected cells, Gpol relocalized from a diffuse distribution in the cytoplasm to punctate foci that colocalized with markers for replication complexes. Expression of Gpol deletion mutants established that the conserved enzymatic domains of Pol were dispensable for replication complex association, but a 38-amino-acid domain in the RdRp unique region of Pol was required. This study demonstrates that viral or virus-induced factors are necessary for Pol to associate with membranes of replication complexes, and it identifies a defined region of Pol that may mediate its interactions with those factors.
Figures





References
-
- Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. - PubMed
-
- Boursnell, M. E., T. D. Brown, I. J. Foulds, P. F. Green, F. M. Tomley, and M. M. Binns. 1987. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J. Gen. Virol. 68:57-77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

