Cell-derived sequences in the N-terminal region of the polyprotein of a cytopathogenic pestivirus
- PMID: 12970452
- PMCID: PMC228520
- DOI: 10.1128/jvi.77.19.10663-10669.2003
Cell-derived sequences in the N-terminal region of the polyprotein of a cytopathogenic pestivirus
Abstract
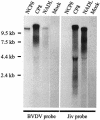
Efficient proteolytic release of nonstructural protein 3 (NS3) from the viral polyprotein is considered to be crucial for the cytopathogenicity of pestiviruses. Here we describe a novel cytopathogenic (cp) bovine viral diarrhea virus strain (BVDV CP8) with a complex insertion composed of viral and cell-derived sequences, including two fragments of the cellular J-domain protein Jiv (J-domain protein interacting with viral protein) located in the N-terminal region of the polyprotein. BVDV CP8 expresses a Jiv fusion protein of 513 amino acids in addition to a complete set of viral proteins. This protein has the capacity to induce NS2-3 cleavage in trans. Accordingly, CP8 is a representative of a novel type of cp pestivirus with a cp-specific mutation located outside of the NS2-3 gene.
Figures




Similar articles
-
Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus.J Virol. 2004 Oct;78(19):10765-75. doi: 10.1128/JVI.78.19.10765-10775.2004. J Virol. 2004. PMID: 15367643 Free PMC article.
-
Cellular sequences in pestivirus genomes encoding gamma-aminobutyric acid (A) receptor-associated protein and Golgi-associated ATPase enhancer of 16 kilodaltons.J Virol. 2002 Dec;76(24):13069-76. doi: 10.1128/jvi.76.24.13069-13076.2002. J Virol. 2002. PMID: 12438634 Free PMC article.
-
Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle.J Virol. 1998 May;72(5):4139-48. doi: 10.1128/JVI.72.5.4139-4148.1998. J Virol. 1998. PMID: 9557703 Free PMC article.
-
Insertion of cellular sequences in the genome of bovine viral diarrhea virus.Arch Virol Suppl. 1991;3:133-42. doi: 10.1007/978-3-7091-9153-8_15. Arch Virol Suppl. 1991. PMID: 9210934 Review.
-
Viral precursor polyproteins: keys of regulation from replication to maturation.Curr Opin Virol. 2013 Apr;3(2):137-42. doi: 10.1016/j.coviro.2013.03.009. Epub 2013 Apr 18. Curr Opin Virol. 2013. PMID: 23602469 Free PMC article. Review.
Cited by
-
Complete genome sequences of a cytopathic/noncytopathic pair of border disease viruses.Genome Announc. 2014 Apr 10;2(2):e00260-14. doi: 10.1128/genomeA.00260-14. Genome Announc. 2014. PMID: 24723711 Free PMC article.
-
CRISPR/Cas9-Mediated Knockout of DNAJC14 Verifies This Chaperone as a Pivotal Host Factor for RNA Replication of Pestiviruses.J Virol. 2019 Feb 19;93(5):e01714-18. doi: 10.1128/JVI.01714-18. Print 2019 Mar 1. J Virol. 2019. PMID: 30518653 Free PMC article.
-
Cytopathogenicity of classical Swine Fever virus correlates with attenuation in the natural host.J Virol. 2008 Oct;82(19):9717-29. doi: 10.1128/JVI.00782-08. Epub 2008 Jul 23. J Virol. 2008. PMID: 18653456 Free PMC article.
-
Cytopathic bovine viral diarrhea viruses (BVDV): emerging pestiviruses doomed to extinction.Vet Res. 2010 Nov-Dec;41(6):44. doi: 10.1051/vetres/2010016. Epub 2010 Mar 4. Vet Res. 2010. PMID: 20197026 Free PMC article. Review.
-
A 45-nucleotide insertion in the NS2 gene is responsible for the cytopathogenicity of a bovine viral diarrhoea virus strain.Virus Genes. 2005 Oct;31(2):135-44. doi: 10.1007/s11262-005-1785-y. Virus Genes. 2005. PMID: 16025238
References
-
- Baroth, M., M. Orlich, H.-J. Thiel, and P. Becher. 2000. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology 278:456-466. - PubMed
-
- Choong, M. L., L. K. Tan, S. L. Lo, E. C. Ren, K. Ou, S. E. Ong, R. C. Liang, T. K. Seow, and M. C. Chung. 2001. An integrated approach in the discovery and characterization of a novel nuclear protein over-expressed in liver and pancreatic tumors. FEBS Lett. 496:109-116. - PubMed
-
- Collett, M. S., R. Larson, C. Gold, D. Strick, D. K. Anderson, and A. F. Purchio. 1988. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology 165:191-199. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

