A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus
- PMID: 12970462
- PMCID: PMC228431
- DOI: 10.1128/jvi.77.19.10725-10729.2003
A lipase isolated from the silkworm Bombyx mori shows antiviral activity against nucleopolyhedrovirus
Abstract
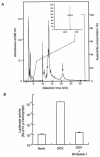
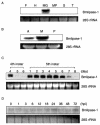
A protein showing strong antiviral activity against Bombyx mori nucleopolyhedrovirus (BmNPV) was purified from the digestive juice of B. mori larvae. A homology search of the deduced amino acid sequence of the protein cDNA revealed 56% homology with Drosophila melanogaster lipase and 21% homology with human lipase. As lipase activity of the protein was confirmed in vitro, this protein was designated Bmlipase-1. Northern blot analysis showed that the Bmlipase-1 gene is expressed in the midgut but not in other tissues, nor is it activated by BmNPV infection. In addition, the Bmlipase-1 gene was shown not to be expressed in the molting and wandering stages, indicating that the gene is hormonally regulated. Our results suggest that an insect digestive enzyme has potential as a physiological barrier against BmNPV at the initial site of viral infection.
Figures



References
-
- Adams, M. D. A. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. - PubMed
-
- Arreguin-Espinosa, R., B. Arreguin, and C. González. 2000. Purification and properties of a lipase from Cephaloleia presignis (Coleoptera, chrysomelidae). Biotechnol. Appl. Biochem. 31:239-244. - PubMed
-
- Barrett, J. W., A. J. Brownwright, M. J. Primavera, A. Retnakaran, and S. R. Palli. 1998. Concomitant primary infection of the midgut epithelial cells and the hemocytes of Trichoplusia ni by Autographa californica nucleopolyhedrovirus. Tissue Cell 30:602-616. - PubMed
-
- Boman, H. G., and D. Hultmark. 1987. Cell-free immunity in insects. Annu. Rev. Microbiol. 41:103-126. - PubMed
-
- Clem, R. J., M. Fechheimer, and L. K. Miller. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388-1390. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
