The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway
- PMID: 12970478
- PMCID: PMC196584
- DOI: 10.1104/pp.103.023358
The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway
Abstract
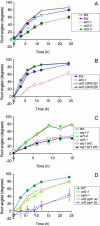
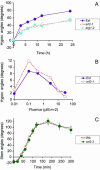
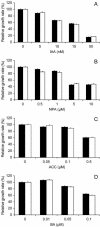
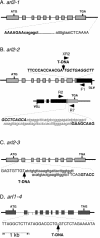
The arl2 mutants of Arabidopsis display altered root and hypocotyl gravitropism, whereas their inflorescence stems are fully gravitropic. Interestingly, mutant roots respond like the wild type to phytohormones and an inhibitor of polar auxin transport. Also, their cap columella cells accumulate starch similarly to wild-type cells, and mutant hypocotyls display strong phototropic responses to lateral light stimulation. The ARL2 gene encodes a DnaJ-like protein similar to ARG1, another protein previously implicated in gravity signal transduction in Arabidopsis seedlings. ARL2 is expressed at low levels in all organs of seedlings and plants. arl2-1 arg1-2 double mutant roots display kinetics of gravitropism similar to those of single mutants. However, double mutants carrying both arl2-1 and pgm-1 (a mutation in the starch-biosynthetic gene PHOSPHOGLUCOMUTASE) at the homozygous state display a more pronounced root gravitropic defect than the single mutants. On the other hand, seedlings with a null mutation in ARL1, a paralog of ARG1 and ARL2, behave similarly to the wild type in gravitropism and other related assays. Taken together, the results suggest that ARG1 and ARL2 function in the same gravity signal transduction pathway in the hypocotyl and root of Arabidopsis seedlings, distinct from the pathway involving PGM.
Figures


References
-
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-814 - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current Protocols in Molecular Biology: Updates 1994-2002. John Wiley and Sons, New York.
-
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137-144 - PubMed
-
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948-950 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

