Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis
- PMID: 12970489
- PMCID: PMC196600
- DOI: 10.1104/pp.103.021089
Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis
Abstract
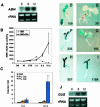
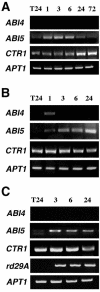
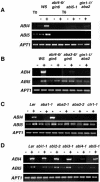
Mutant characterization has demonstrated that ABI4 (Abscisic Acid [ABA] Insensitive 4), ABI5 (ABA Insensitive 5), and CTR1 (Constitutive Triple Response 1) genes play an important role in the sugar signaling response in plants. The present study shows that the transcripts of these three genes are modulated by glucose (Glc) independently of the developmental arrest caused by high Glc concentrations. ABI4 and ABI5 transcripts accumulate in response to sugars, whereas the CTR1 transcript is transiently reduced followed by a rapid recovery. The results of our kinetic studies on gene expression indicate that ABI4, ABI5, and CTR1 are regulated by multiple signals including Glc, osmotic stress, and ABA. However, the differential expression profiles caused by these treatments suggest that distinct signaling pathways are used for each signal. ABI4 and ABI5 response to the Glc analog 2-deoxy-Glc supports this conclusion. Glc regulation of ABI4 and CTR1 transcripts is dependent on the developmental stage. Finally, the Glc-mediated regulation of ABI4 and ABI5 is affected in mutants displaying Glc-insensitive phenotypes such as gins, abas, abi4, abi5, and ctr1 but not in abi1-1, abi2-1, and abi3-1, which do not show a Glc-insensitive phenotype. The capacity of transcription factors, like the ones analyzed in this work, to be regulated by a variety of signals might contribute to the ability of plants to respond in a flexible and integral way to continuous changes in the internal and external environment.
Figures


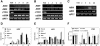
References
-
- Arroyo A, Arenas F, Jiménez A, Cantero A, León P (2001) The participation of ABA in the glucose-mediated regulation in Arabidopsis. Presented at 11th International Conference on Arabidopsis Research, June 23-27, 2001, Madison, WI
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Siedman JG, Smith JA, Struhl K (1989) Current Protocols in Molecular Biology. John Wiley & Sons, New York
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

