Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different European origins: a single shared haplotype indicates an ancestral founder effect
- PMID: 12970845
- PMCID: PMC1180606
- DOI: 10.1086/378566
Confirmation of the type 2 myotonic dystrophy (CCTG)n expansion mutation in patients with proximal myotonic myopathy/proximal myotonic dystrophy of different European origins: a single shared haplotype indicates an ancestral founder effect
Abstract
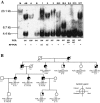
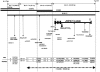
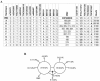
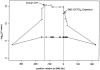
Myotonic dystrophy (DM), the most common form of muscular dystrophy in adults, is a clinically and genetically heterogeneous neuromuscular disorder. DM is characterized by autosomal dominant inheritance, muscular dystrophy, myotonia, and multisystem involvement. Type 1 DM (DM1) is caused by a (CTG)(n) expansion in the 3' untranslated region of DMPK in 19q13.3. Multiple families, predominantly of German descent and with clinically variable presentation that included proximal myotonic myopathy (PROMM) and type 2 DM (DM2) but without the DM1 mutation, showed linkage to the 3q21 region and were recently shown to segregate a (CCTG)(n) expansion mutation in intron 1 of ZNF9. Here, we present linkage to 3q21 and mutational confirmation in 17 kindreds of European origin with PROMM and proximal myotonic dystrophy, from geographically distinct populations. All patients have the DM2 (CCTG)(n) expansion. To study the evolution of this mutation, we constructed a comprehensive physical map of the DM2 region around ZNF9. High-resolution haplotype analysis of disease chromosomes with five microsatellite and 22 single-nucleotide polymorphism markers around the DM2 mutation identified extensive linkage disequilibrium and a single shared haplotype of at least 132 kb among patients from the different populations. With the exception of the (CCTG)(n) expansion, the available markers indicate that the DM2 haplotype is identical to the most common haplotype in normal individuals. This situation is reminiscent of that seen in DM1. Taken together, these data suggest a single founding mutation in DM2 patients of European origin. We estimate the age of the founding haplotype and of the DM2 (CCTG) expansion mutation to be approximately 200-540 generations.
Figures



References
Electronic-Database Information
-
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for RP11-814L21 [accession number AC022944], RP11-309B5 [accession number AC135587], RP11-723O4 [accession number AC112484], and RP11-434H6 [accession number AC108673], and chromosome 3 contig [accession number NT_006025])
-
- Gene Viewer, Cancer Genome Anatomy Project (CGAP), http://gai.nci.nih.gov/cgi-bin/GeneViewer.cgi (for CGAP SNPs)
-
- NCBI Reference Sequence (RefSeq), http://www.ncbi.nlm.nih.gov/RefSeq/
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DM1, DM2, and PROMM)
References
-
- Abbruzzese C, Krahe R, Liguori M, Tessarolo D, Siciliano MJ, Ashizawa T, Giacanelli M (1996) Myotonic dystrophy phenotype without expansion of (CTG)n repeat: an entity distinct from proximal myotonic myopathy (PROMM)? J Neurol 243:715–721 - PubMed
-
- Ashizawa T, Epstein HF (1991) Ethnic distribution of myotonic dystrophy gene. Lancet 338:642–643 - PubMed
-
- Bassez G, Attarian S, Laforet P, Azulay JP, Rouche A, Ferrer X, Urtizberea JA, Pellissier JF, Duboc D, Fardeau M, Pouget J, Eymard B (2001) Proximal myotonial myopathy (PROMM): clinical and histology study. Rev Neurol 157:209–218 - PubMed
-
- Borrego S, Wright FA, Fernandez RM, Williams N, Lopez-Alonso M, Davuluri R, Antinolo G, Eng C (2003) A founding locus within the RET proto-oncogene may account for a large proportion of apparently sporadic Hirschsprung disease and a subset of cases of sporadic medullary thyroid carcinoma. Am J Hum Genet 72:88–100 - PMC - PubMed
-
- Brock GJ, Anderson NH, Monckton DG (1999) Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum Mol Genet 8:1061–1067 - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

