Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding
- PMID: 12972571
- PMCID: PMC196582
- DOI: 10.1091/mbc.e03-02-0090
Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding
Abstract
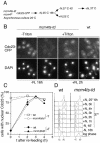
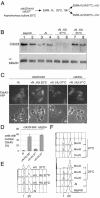
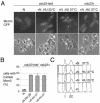
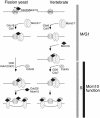
Using a cytological assay to monitor the successive chromatin association of replication proteins leading to replication initiation, we have investigated the function of fission yeast Cdc23/Mcm10 in DNA replication. Inactivation of Cdc23 before replication initiation using tight degron mutations has no effect on Mcm2 chromatin association, and thus pre-replicative complex (pre-RC) formation, although Cdc45 chromatin binding is blocked. Inactivating Cdc23 during an S phase block after Cdc45 has bound causes a small reduction in Cdc45 chromatin binding, and replication does not terminate in the absence of Mcm10 function. These observations show that Cdc23/Mcm10 function is conserved between fission yeast and Xenopus, where in vitro analysis has indicated a similar requirement for Cdc45 binding, but apparently not compared with Saccharomyces cerevisiae, where Mcm10 is needed for Mcm2 chromatin binding. However, unlike the situation in Xenopus, where Mcm10 chromatin binding is dependent on Mcm2-7, we show that the fission yeast protein is bound to chromatin throughout the cell cycle in growing cells, and only displaced from chromatin during quiescence. On return to growth, Cdc23 chromatin binding is rapidly reestablished independently from pre-RC formation, suggesting that chromatin association of Cdc23 provides a link between proliferation and competence to execute DNA replication.
Figures
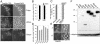




References
-
- Aparicio, O.M., Weinstein, D.M., and Bell, S.P. (1997). Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69. - PubMed
-
- Aves, S.J., Tongue, N., Foster, A.J., and Hart, E.A. (1998). The essential Schizosaccharomyces pombe cdc23 DNA replication gene shares structural and functional homology with the Saccharomyces cerevisiae DNA43 (MCM10) gene. Curr. Genet. 34, 164–171. - PubMed
-
- Basi, G., Schmid, E., and Maundrell, K. (1993). TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123, 131–136. - PubMed
-
- Bell, S.P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev Biochem. 71, 333–374. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

