The yeast hnRNP-like protein Hrp1/Nab4 sccumulates in the cytoplasm after hyperosmotic stress: a novel Fps1-dependent response
- PMID: 12972575
- PMCID: PMC196592
- DOI: 10.1091/mbc.e03-01-0854
The yeast hnRNP-like protein Hrp1/Nab4 sccumulates in the cytoplasm after hyperosmotic stress: a novel Fps1-dependent response
Abstract
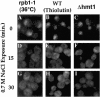
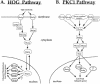
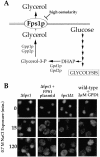
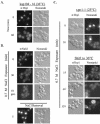
The Hrp1/Nab4 shuttling protein belongs to a family of RNA binding proteins that bind to nascent RNA polymerase II transcripts and form hnRNP complexes. Members of this family function in a staggering array of cellular activities, ranging from transcription and pre-mRNA processing in the nucleus to cytoplasmic mRNA translation and turnover. It has recently been recognized that the yeast stress response can include alterations in hnRNP-mediated mRNA export. We now report that the steady-state localization of Hrp1p rapidly shifts from the nucleus to the cytoplasm in response to osmotic stress. In contrast to a general stress response resulting in a transient relocation, Hrp1p redistribution is specific to hyperosmotic stress and is only reversed after stress removal. Hrp1p relocalization requires both the CRM1/XPO1 exportin and the FPS1 glycerol transporter genes but is independent of ongoing RNA transcription and protein arginine methylation. However, mutations in the high osmolarity glycerol and protein kinase C osmosensing pathways do not impact the Hrp1p hyperosmotic response. We present a working model for the cytoplasmic accumulation of Hrp1 and discuss the implications of this relocalization on Hrp1p function.
Figures





Similar articles
-
Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p.Mol Cell Biol. 2004 Dec;24(24):10742-56. doi: 10.1128/MCB.24.24.10742-10756.2004. Mol Cell Biol. 2004. PMID: 15572678 Free PMC article.
-
The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay.Mol Cell. 2000 Mar;5(3):489-99. doi: 10.1016/s1097-2765(00)80443-8. Mol Cell. 2000. PMID: 10882134
-
Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3'-end formation.RNA. 1999 Feb;5(2):272-80. doi: 10.1017/s1355838299981633. RNA. 1999. PMID: 10024178 Free PMC article.
-
[Quality control of pre-ribosome coupled with ribosome biogenesis].Seikagaku. 2014 Dec;86(6):793-7. Seikagaku. 2014. PMID: 25675820 Review. Japanese. No abstract available.
-
Dbp5 - from nuclear export to translation.Biochim Biophys Acta. 2013 Aug;1829(8):791-8. doi: 10.1016/j.bbagrm.2012.10.010. Epub 2012 Nov 2. Biochim Biophys Acta. 2013. PMID: 23128325 Review.
Cited by
-
Functional specificity of shuttling hnRNPs revealed by genome-wide analysis of their RNA binding profiles.RNA. 2005 Apr;11(4):383-93. doi: 10.1261/rna.7234205. Epub 2005 Feb 9. RNA. 2005. PMID: 15703440 Free PMC article.
-
Insights into the Mechanism of Pre-mRNA Splicing of Tiny Introns from the Genome of a Giant Ciliate Stentor coeruleus.Int J Mol Sci. 2022 Sep 19;23(18):10973. doi: 10.3390/ijms231810973. Int J Mol Sci. 2022. PMID: 36142882 Free PMC article.
-
Domains and residues of the Saccharomyces cerevisiae hnRNP protein Hrp1 important for transcriptional autoregulation and noncoding RNA termination.Genetics. 2023 Aug 31;225(1):iyad134. doi: 10.1093/genetics/iyad134. Genetics. 2023. PMID: 37467478 Free PMC article.
-
The hnRNP-like Nab3 termination factor can employ heterologous prion-like domains in place of its own essential low complexity domain.PLoS One. 2017 Oct 12;12(10):e0186187. doi: 10.1371/journal.pone.0186187. eCollection 2017. PLoS One. 2017. PMID: 29023495 Free PMC article.
-
Noncanonical transcript forms in yeast and their regulation during environmental stress.RNA. 2010 Jun;16(6):1256-67. doi: 10.1261/rna.2038810. Epub 2010 Apr 26. RNA. 2010. PMID: 20421314 Free PMC article.
References
-
- Aitchison, J.D., Blobel, G., and Rout, M.P. (1996). Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274, 624–627. - PubMed
-
- Arts, G.J., Fornerod, M., and Mattaj, I.W. (1998). Identification of a nuclear export receptor for tRNA. Curr. Biol. 8, 305–314. - PubMed
-
- Boeke, J.D., LaCroute, F., and Fink, G.R. (1984). A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. PG. Mol. Gen. Genet. 197, 345–347. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

