Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse
- PMID: 12972605
- PMCID: PMC193944
- DOI: 10.1128/MCB.23.19.6857-6875.2003
Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse
Abstract
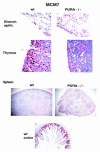
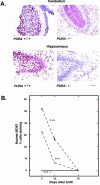
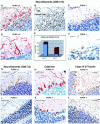
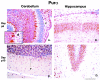
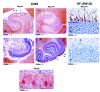
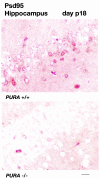
The single-stranded DNA- and RNA-binding protein, Puralpha, has been implicated in many biological processes, including control of transcription of multiple genes, initiation of DNA replication, and RNA transport and translation. Deletions of the PURA gene are frequent in acute myeloid leukemia. Mice with targeted disruption of the PURA gene in both alleles appear normal at birth, but at 2 weeks of age, they develop neurological problems manifest by severe tremor and spontaneous seizures and they die by 4 weeks. There are severely lower numbers of neurons in regions of the hippocampus and cerebellum of PURA(-/-) mice versus those of age-matched +/+ littermates, and lamination of these regions is aberrant at time of death. Immunohistochemical analysis of MCM7, a protein marker for DNA replication, reveals a lack of proliferation of precursor cells in these regions in the PURA(-/-) mice. Levels of proliferation were also absent or low in several other tissues of the PURA(-/-) mice, including those of myeloid lineage, whereas those of PURA(+/-) mice were intermediate. Evaluation of brain sections indicates a reduction in myelin and glial fibrillary acidic protein labeling in oligodendrocytes and astrocytes, respectively, indicating pathological development of these cells. At postnatal day 5, a critical time for cerebellar development, Puralpha and Cdk5 were both at peak levels in bodies and dendrites of Purkinje cells of PURA(+/+) mice, but both were absent in dendrites of PURA(-/-) mice. Puralpha and Cdk5 can be coimmunoprecipitated from brain lysates of PURA(+/+) mice. Immunohistochemical studies reveal a dramatic reduction in the level of both phosphorylated and nonphosphorylated neurofilaments in dendrites of the Purkinje cell layer and of synapse formation in the hippocampus. Overall results are consistent with a role for Puralpha in developmentally timed DNA replication in specific cell types and also point to a newly emerging role in compartmentalized RNA transport and translation in neuronal dendrites.
Figures





References
-
- Barr, S. M., and E. M. Johnson. 2001. Ras-induced colony formation and anchorage-independent growth inhibited by elevated expression of Puralpha in NIH3T3 cells. J. Cell. Biochem. 81:621-638. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
