Autoregulation of mouse histone deacetylase 1 expression
- PMID: 12972616
- PMCID: PMC193942
- DOI: 10.1128/MCB.23.19.6993-7004.2003
Autoregulation of mouse histone deacetylase 1 expression
Abstract
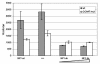
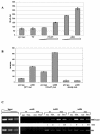
Histone deacetylase 1 (HDAC1) is a major regulator of chromatin structure and gene expression. Tight control of HDAC1 expression is essential for development and normal cell cycle progression. In this report, we analyzed the regulation of the mouse HDAC1 gene by deacetylases and acetyltransferases. The murine HDAC1 promoter lacks a TATA box consensus sequence but contains several putative SP1 binding sites and a CCAAT box, which is recognized by the transcription factor NF-Y. HDAC1 promoter-reporter studies revealed that the distal SP1 site and the CCAAT box are crucial for HDAC1 promoter activity and act synergistically to constitute HDAC1 promoter activity. Furthermore, these sites are essential for activation of the HDAC1 promoter by the deacetylase inhibitor trichostatin A (TSA). Chromatin immunoprecipitation assays showed that HDAC1 is recruited to the promoter by SP1 and NF-Y, thereby regulating its own expression. Coexpression of acetyltransferases elevates HDAC1 promoter activity when the SP1 site and the CCAAT box are intact. Increased histone acetylation at the HDAC1 promoter region in response to TSA treatment is dependent on binding sites for SP1 and NF-Y. Taken together, our results demonstrate for the first time the autoregulation of a histone-modifying enzyme in mammalian cells.
Figures







Similar articles
-
The histone deacetylase inhibitor trichostatin A derepresses the telomerase reverse transcriptase (hTERT) gene in human cells.Exp Cell Res. 2002 Mar 10;274(1):25-34. doi: 10.1006/excr.2001.5462. Exp Cell Res. 2002. PMID: 11855854
-
Histone deacetylase inhibitors stimulate mitochondrial HMG-CoA synthase gene expression via a promoter proximal Sp1 site.Nucleic Acids Res. 2003 Mar 15;31(6):1693-703. doi: 10.1093/nar/gkg262. Nucleic Acids Res. 2003. PMID: 12626711 Free PMC article.
-
Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex.J Biol Chem. 2002 Sep 6;277(36):33431-8. doi: 10.1074/jbc.M204417200. Epub 2002 Jun 28. J Biol Chem. 2002. PMID: 12091390
-
Histone deacetylase inhibitor activates the p21/WAF1/Cip1 gene promoter through the Sp1 sites.Ann N Y Acad Sci. 1999;886:195-9. doi: 10.1111/j.1749-6632.1999.tb09415.x. Ann N Y Acad Sci. 1999. PMID: 10667218 Review.
-
HDAC1: an environmental sensor regulating endothelial function.Cardiovasc Res. 2022 Jun 29;118(8):1885-1903. doi: 10.1093/cvr/cvab198. Cardiovasc Res. 2022. PMID: 34264338 Free PMC article. Review.
Cited by
-
Correlation of epigenetic aberrance with STAT3 signaling pathway in gastric carcinogenesis.Dig Dis Sci. 2012 Aug;57(8):2055-62. doi: 10.1007/s10620-012-2152-1. Epub 2012 May 6. Dig Dis Sci. 2012. PMID: 22562532
-
Mechanism, Consequences, and Therapeutic Targeting of Abnormal IL15 Signaling in Cutaneous T-cell Lymphoma.Cancer Discov. 2016 Sep;6(9):986-1005. doi: 10.1158/2159-8290.CD-15-1297. Epub 2016 Jul 15. Cancer Discov. 2016. PMID: 27422033 Free PMC article.
-
Promoter conservation in HDACs points to functional implications.BMC Genomics. 2019 Jul 27;20(1):613. doi: 10.1186/s12864-019-5973-x. BMC Genomics. 2019. PMID: 31351464 Free PMC article.
-
Patho-epigenetics: histone deacetylases as targets of pathogens and therapeutics.Microlife. 2021 Nov 29;2:uqab013. doi: 10.1093/femsml/uqab013. eCollection 2021. Microlife. 2021. PMID: 37223249 Free PMC article.
-
NF-Y behaves as a bifunctional transcription factor that can stimulate or repress the FGF-4 promoter in an enhancer-dependent manner.Gene Expr. 2005;12(3):193-212. doi: 10.3727/000000005783992052. Gene Expr. 2005. PMID: 16128003 Free PMC article.
References
-
- Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. - PubMed
-
- Baker, A., M. Saltik, H. Lehrmann, I. Killisch, V. Mautner, G. Lamm, G. Christofori, and M. Cotten. 1997. Polyethylenimine (PEI) is a simple, inexpensive and effective reagent for condensing and linking plasmid DNA to adenovirus for gene delivery. Gene Ther. 4:773-782. - PubMed
-
- Billon, N., D. Carlisi, M. B. Datto, L. A. van Grunsven, A. Watt, X. F. Wang, and B. B. Rudkin. 1999. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 18:2872-2882. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
