Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein
- PMID: 12972647
- PMCID: PMC219055
- DOI: 10.1104/pp.103.027870
Characterization of the targeting signal of the Arabidopsis 22-kD integral peroxisomal membrane protein
Abstract
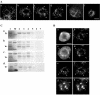
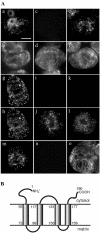
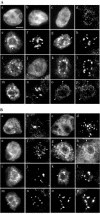
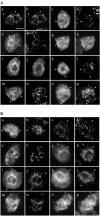
Using a combination of in vivo and in vitro assays, we characterized the sorting pathway and molecular targeting signal for the Arabidopsis 22-kD peroxisome membrane protein (PMP22), an integral component of the membrane of all peroxisomes in the mature plant. We show that nascent PMP22 is sorted directly from the cytosol to peroxisomes and that it is inserted into the peroxisomal boundary membrane with its N- and C-termini facing the cytosol. This direct sorting of PMP22 to peroxisomes contrasts with the indirect sorting reported previously for cottonseed (Gossypium hirsutum) ascorbate peroxidase, an integral PMP that sorts to peroxisomes via a subdomain of the endoplasmic reticulum. Thus, at least two different sorting pathways for PMPs exist in plant cells. At least four distinct regions within the N-terminal one-half of PMP22, including a positively charged domain present in most peroxisomal integral membrane-destined proteins, functions in a cooperative manner in efficient peroxisomal targeting and insertion. In addition, targeting with high fidelity to peroxisomes requires all four membrane-spanning domains in PMP22. Together, these results illustrate that the PMP22 membrane peroxisomal targeting signal is complex and that different elements within the signal may be responsible for mediating unique aspects of PMP22 biogenesis, including maintaining the solubility before membrane insertion, targeting to peroxisomes, and ensuring proper assembly in the peroxisomal boundary membrane.
Figures



Similar articles
-
Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S.cerevisiae, causes proliferation of the endoplasmic reticulum membrane.EMBO J. 1997 Dec 15;16(24):7326-41. doi: 10.1093/emboj/16.24.7326. EMBO J. 1997. PMID: 9405362 Free PMC article.
-
The peroxisomal membrane targeting elements of human peroxin 2 (PEX2).Eur J Cell Biol. 2003 Apr;82(4):155-62. doi: 10.1078/0171-9335-00310. Eur J Cell Biol. 2003. PMID: 12751901
-
Arabidopsis peroxisomes possess functionally redundant membrane and matrix isoforms of monodehydroascorbate reductase.Plant J. 2005 Sep;43(6):900-14. doi: 10.1111/j.1365-313X.2005.02503.x. Plant J. 2005. PMID: 16146528
-
Targeting and insertion of peroxisomal membrane proteins: ER trafficking versus direct delivery to peroxisomes.Biochim Biophys Acta. 2016 May;1863(5):870-80. doi: 10.1016/j.bbamcr.2015.09.021. Epub 2015 Sep 25. Biochim Biophys Acta. 2016. PMID: 26392202 Review.
-
Targeting signals in peroxisomal membrane proteins.Biochim Biophys Acta. 2006 Dec;1763(12):1629-38. doi: 10.1016/j.bbamcr.2006.08.020. Epub 2006 Aug 25. Biochim Biophys Acta. 2006. PMID: 17020786 Review.
Cited by
-
Go your own way: membrane-targeting sequences.Plant Physiol. 2021 Apr 2;185(3):608-618. doi: 10.1093/plphys/kiaa058. Plant Physiol. 2021. PMID: 33822216 Free PMC article.
-
Peroxisome biogenesis and the role of protein import.J Cell Mol Med. 2003 Oct-Dec;7(4):388-400. doi: 10.1111/j.1582-4934.2003.tb00241.x. J Cell Mol Med. 2003. PMID: 14754507 Free PMC article. Review.
-
Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes.Plant Cell. 2008 Jun;20(6):1567-85. doi: 10.1105/tpc.107.057679. Epub 2008 Jun 6. Plant Cell. 2008. PMID: 18539750 Free PMC article.
-
Defining the plant peroxisomal proteome: from Arabidopsis to rice.Front Plant Sci. 2011 Dec 27;2:103. doi: 10.3389/fpls.2011.00103. eCollection 2011. Front Plant Sci. 2011. PMID: 22645559 Free PMC article.
-
Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting.J Cell Biol. 2008 Jan 28;180(2):325-39. doi: 10.1083/jcb.200705049. J Cell Biol. 2008. PMID: 18227279 Free PMC article.
References
-
- Baerends RJ, Faber KN, Kram AM, Kiel JAKW, van der Klei IJ, Veenhuis M (2000) A stretch of positively charged amino acids at the N terminus of Hansenula polymorpha Pex3p is involved in incorporation of the protein into the peroxisomal membrane. J Biol Chem 275: 9985–9995 - PubMed
-
- Brosius U, Dehmel T, Gärtner J (2002) Two different targeting signals direct human peroxisomal membrane protein 22 to peroxisomes. J Biol Chem 277: 774–784 - PubMed
-
- Corpas FJ, Barroso JB, del Rio LA (2001) Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells. Trends Plant Sci 6: 145–150 - PubMed
-
- Coughlan SJ, Hastings C, Winfrey R (1997) Congin and characterization of the calreticulin gene from Ricinus communis L. Plant Mol Biol 34: 897–911 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

