Isolation of a gene encoding a copper chaperone for the copper/zinc superoxide dismutase and characterization of its promoter in potato
- PMID: 12972661
- PMCID: PMC219038
- DOI: 10.1104/pp.103.025320
Isolation of a gene encoding a copper chaperone for the copper/zinc superoxide dismutase and characterization of its promoter in potato
Abstract
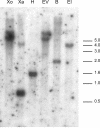
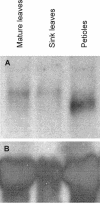
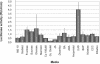
Gene expression during the potato (Solanum tuberosum) tuber lifecycle was monitored by cDNA-amplified fragment-length polymorphism, and several differentially expressed transcript-derived fragments were isolated. One fragment, named TDFL431, showed high homology to a copper (Cu) chaperone for Cu/zinc superoxide dismutase (CCS). The Ccs protein is responsible for the delivery of Cu to the Cu/zinc superoxide dismutase enzyme. The potato CCS (StCCS) full-length gene was isolated, and its sequence was compared with CCSs from other species. The promoter region of this gene was isolated, fused to the firefly luciferase coding sequence, and used for transformation of potato plants. The highest level of StCCS-luciferase expression was detected in the cortex of stem (like) tissues, such as stem nodes, stolons, and tubers; lower levels were detected in roots and flowers. The StCCS promoter contains regions highly homologous to several plant cis-acting elements. Three of them are related to auxin response, whereas four others are related to response to various stresses. Induction of the StCCS promoter was analyzed on 18 media, differing in hormone, sugar, and Cu content. StCCS expression was induced by auxin, gibberellins (GA4 + 7), fructose, sucrose, and glucose and was inhibited by relatively high concentrations of Cu.
Figures



Similar articles
-
Molecular Characterization of MaCCS, a Novel Copper Chaperone Gene Involved in Abiotic and Hormonal Stress Responses in Musa acuminata cv. Tianbaojiao.Int J Mol Sci. 2016 Mar 24;17(4):441. doi: 10.3390/ijms17040441. Int J Mol Sci. 2016. PMID: 27023517 Free PMC article.
-
Cloning and characterization of a cDNA encoding the cytosolic copper/zinc-superoxide dismutase from sweet potato tuberous root.Plant Mol Biol. 1993 Nov;23(4):911-3. doi: 10.1007/BF00021547. Plant Mol Biol. 1993. PMID: 8251645
-
Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: regulation and unexpected phenotypes in an Arabidopsis mutant.Mol Plant. 2009 Nov;2(6):1336-50. doi: 10.1093/mp/ssp084. Epub 2009 Oct 21. Mol Plant. 2009. PMID: 19969519
-
Molecular characterization of StCDPK1, a calcium-dependent protein kinase from Solanum tuberosum that is induced at the onset of tuber development.Plant Mol Biol. 2001 Jul;46(5):591-601. doi: 10.1023/a:1010661304980. Plant Mol Biol. 2001. PMID: 11516152
-
Evolutionary aspects of superoxide dismutase: the copper/zinc enzyme.Free Radic Res Commun. 1991;12-13 Pt 1:349-61. doi: 10.3109/10715769109145804. Free Radic Res Commun. 1991. PMID: 2071039 Review.
Cited by
-
Computational exploration of cis-regulatory modules in rhythmic expression data using the "Exploration of Distinctive CREs and CRMs" (EDCC) and "CRM Network Generator" (CNG) programs.PLoS One. 2018 Jan 3;13(1):e0190421. doi: 10.1371/journal.pone.0190421. eCollection 2018. PLoS One. 2018. PMID: 29298348 Free PMC article.
-
A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis.Plant Physiol. 2005 Sep;139(1):425-36. doi: 10.1104/pp.105.065284. Epub 2005 Aug 26. Plant Physiol. 2005. PMID: 16126858 Free PMC article.
-
Molecular Characterization of MaCCS, a Novel Copper Chaperone Gene Involved in Abiotic and Hormonal Stress Responses in Musa acuminata cv. Tianbaojiao.Int J Mol Sci. 2016 Mar 24;17(4):441. doi: 10.3390/ijms17040441. Int J Mol Sci. 2016. PMID: 27023517 Free PMC article.
References
-
- Allen RD, Webb RP, Schake SA (1997) Use of transgenic plants to study antioxidant defenses. Free Radic Biol Med 23: 473–479 - PubMed
-
- Asada K (1992) Ascorbate peroxidase: a hydrogen peroxide-scavenging enzyme in plants. Physiol Plant 85: 235–241
-
- Bachem C, van der Hoeven R, Lucker J, Oomen R, Casarini E, Jacobsen E, Visser R (2000) Functional genomic analysis of potato tuber life-cycle. Potato Res 43: 297–312
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

