Helicases as antiviral drug targets
- PMID: 12973446
- PMCID: PMC3571683
- DOI: 10.1358/dnp.2003.16.6.829307
Helicases as antiviral drug targets
Abstract
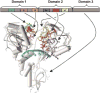
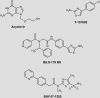
Helicases catalytically unwind duplex DNA or RNA using energy derived from the hydrolysis of nucleoside triphosphates and are attractive drug targets because they are required for viral replication. This review discusses methods for helicase identification, classification and analysis, and presents an overview of helicases that are necessary for the replication of human pathogenic viruses. Newly developed methods to analyze helicases, coupled with recently determined atomic structures, have led to a better understanding of their mechanisms of action. The majority of this research has concentrated on enzymes encoded by the herpes simplex virus (HSV) and the hepatitis C virus (HCV). Helicase inhibitors that target the HSV helicase-primase complex comprised of the UL5, UL8 and UL52 proteins have recently been shown to effectively control HSV infection in animal models. In addition, several groups have reported structures of the HCV NS3 helicase at atomic resolutions, and mechanistic studies have uncovered characteristics that distinguish the HCV helicase from related cellular proteins. These new developments should eventually lead to new antiviral medications.
(c) 2003 Prous Science. All rights reserved.
Figures

References
-
- Nakayama H. RecQ family helicases: Roles as tumor suppressor proteins. Oncogene. 2002;21:9008–21. - PubMed
-
- Crumpacker CS, Schaffer PA. New anti-HSV therapeutics target the helicase- primase complex. Nat Med. 2002;8:327–8. - PubMed
-
- Wong I, Moore KJ, Bjornson KP, Hsieh J, Lohman TM. ATPase activity of Escherichia coli Rep helicase is dramatically dependent on DNA ligation and protein oligomeric states. Biochemistry. 1996;35:5726–34. - PubMed
-
- Preugschat F, Averett DR, Clarke BE, Porter DJT. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J Biol Chem. 1996;271:24449–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
