Adenoviral vectors expressing siRNAs for discovery and validation of gene function
- PMID: 12975310
- PMCID: PMC403715
- DOI: 10.1101/gr.1332603
Adenoviral vectors expressing siRNAs for discovery and validation of gene function
Abstract
RNA interference is a powerful tool for studying gene function and for drug target discovery in diverse organisms and cell types. In mammalian systems, small interfering RNAs (siRNAs), or DNA plasmids expressing these siRNAs, have been used to down-modulate gene expression. However, inefficient transfection protocols, in particular, for primary cell types, have hampered the use of these tools in disease-relevant cellular assays. To be able to use this technology for genome-wide function screening, a more robust transduction protocol, resulting in a longer duration of the knock-down effect, is required. Here, we describe the validation of adenoviral vectors that express hairpin RNAs that are further processed to siRNAs. Infection of cell lines, or primary human cells, with these viruses leads to an efficient, sequence-specific, and prolonged reduction of the corresponding target mRNA, resulting in a reduction of the encoded protein level in the cell. For knock-down of one of the targets, GalphaS, we have measured inhibition of ligand-dependent, G-protein-coupled signaling. It is expected that this technology will prove to be of great value in target validation and target discovery efforts.
Figures

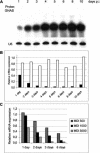

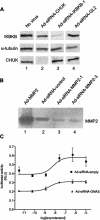
References
-
- Ashrafi, K., Chang, F.Y., Watts, J.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268-272. - PubMed
-
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002a. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550-553. - PubMed
-
- ____. 2002b. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243-247. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
