Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation
- PMID: 12975311
- PMCID: PMC403708
- DOI: 10.1101/gr.1349003
Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation
Abstract
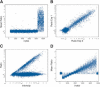
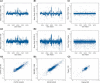
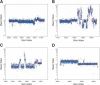
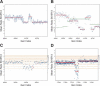
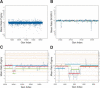
We have developed a methodology we call ROMA (representational oligonucleotide microarray analysis), for the detection of the genomic aberrations in cancer and normal humans. By arraying oligonucleotide probes designed from the human genome sequence, and hybridizing with "representations" from cancer and normal cells, we detect regions of the genome with altered "copy number." We achieve an average resolution of 30 kb throughout the genome, and resolutions as high as a probe every 15 kb are practical. We illustrate the characteristics of probes on the array and accuracy of measurements obtained using ROMA. Using this methodology, we identify variation between cancer and normal genomes, as well as between normal human genomes. In cancer genomes, we readily detect amplifications and large and small homozygous and hemizygous deletions. Between normal human genomes, we frequently detect large (100 kb to 1 Mb) deletions or duplications. Many of these changes encompass known genes. ROMA will assist in the discovery of genes and markers important in cancer, and the discovery of loci that may be important in inherited predispositions to disease.
Figures

References
-
- Bartlett, J. and Mallon, E.C.T. 2003. The clinical evaluation of HER-2 status: Which test to use? J. Pathology 199: 418-423. - PubMed
-
- Chee, M., Yang, R., Hubbell, E., Berno, A., Huang, X.C., Stern, D., Winkler, J., Lockhard, D.J., Morris, M.S., and Fodor, S.P. 1996. Accessing genetic information with high-density DNA arrays. Science 274: 610-614. - PubMed
-
- DeRisi, J., Penland, L., Brown, P.O., Bittner, M.L., Meltzer, P.S., Ray, M., Chen, Y., Su, Y.A., and Trent, J.M. 1996. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 14: 457-460. - PubMed
-
- Golub, T.R., Slonim, D.K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J.P., Coller, H., Loh, M.L., Downing, J.R., Caligiuri, M.A., et al. 1999. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science 286: 531-537. - PubMed
WEB SITE REFERENCES
-
- http://roma.cshl.org/; ROMA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
