Phage N4 RNA polymerase II recruitment to DNA by a single-stranded DNA-binding protein
- PMID: 12975320
- PMCID: PMC196469
- DOI: 10.1101/gad.1121403
Phage N4 RNA polymerase II recruitment to DNA by a single-stranded DNA-binding protein
Abstract
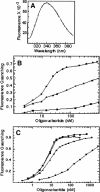
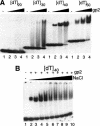
Transcription of bacteriophage N4 middle genes is carried out by a phage-coded, heterodimeric RNA polymerase (N4 RNAPII), which belongs to the family of T7-like RNA polymerases. In contrast to phage T7-RNAP, N4 RNAPII displays no activity on double-stranded templates and low activity on single-stranded templates. In vivo, at least one additional N4-coded protein (p17) is required for N4 middle transcription. We show that N4 ORF2 encodes p17 (gp2). Characterization of purified gp2revealed that it is a single-stranded DNA-binding protein that activates N4 RNAPII transcription on single-stranded DNA templates through specific interaction with N4 RNAPII. On the basis of the properties of the proteins involved in N4 RNAPII transcription and of middle promoters, we propose a model for N4 RNAPII promoter recognition, in which gp2plays two roles, stabilization of a single-stranded region at the promoter and recruitment of N4 RNAPII through gp2-N4 RNAPII interactions. Furthermore, we discuss our results in the context of transcription initiation by mitochondrial RNA polymerases.
Figures
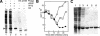

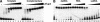

References
-
- Abravaya K. and Rothman-Denes, L.B. 1989a. In vitro requirements for N4 RNA polymerase II-specific initiation. J. Biol. Chem. 264: 12695-12699. - PubMed
-
- ____. 1989b. N4 RNA polymerase II sites of transcription initiation. J. Mol. Biol. 211: 359-372. - PubMed
-
- Ausubel F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (eds.) 1999. Current protocols in molecular biology. Wiley, New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
