Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers
- PMID: 12975468
- PMCID: PMC193667
- DOI: 10.1172/JCI18509
Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers
Abstract
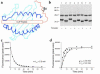
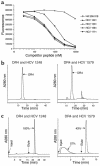
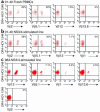
Containment of hepatitis C virus (HCV) and other chronic human viral infections is associated with persistence of virus-specific CD4 T cells, but ex vivo characterization of circulating CD4 T cells has not been achieved. To further define the phenotype and function of these cells, we developed a novel approach for the generation of tetrameric forms of MHC class II/peptide complexes that is based on the cellular peptide-exchange mechanism. HLA-DR molecules were expressed as precursors with a covalently linked CLIP peptide, which could be efficiently exchanged with viral peptides following linker cleavage. In subjects who spontaneously resolved HCV viremia, but not in those with chronic progressive infection, HCV tetramer-labeled cells could be isolated by magnetic bead capture despite very low frequencies (1:1,200 to 1:111,000) among circulating CD4 T cells. These T cells expressed a set of surface receptors (CCR7+CD45RA-CD27+) indicative of a surveillance function for secondary lymphoid structures and had undergone significant in vivo selection since they utilized a restricted Vbeta repertoire. These studies demonstrate a relationship between clinical outcome and the presence of circulating CD4 T cells directed against this virus. Moreover, they show that rare populations of memory CD4 T cells can be studied ex vivo in human diseases.
Figures




References
-
- Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. - PubMed
-
- Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat. Med. 2001;7:913–919. - PubMed
-
- Lauer GM, Walker BD. Hepatitis C virus infection. N. Engl. J. Med. 2001;345:41–52. - PubMed
-
- Alter MJ, et al. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 1999;341:556–562. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

