Efficiency of a programmed -1 ribosomal frameshift in the different subtypes of the human immunodeficiency virus type 1 group M
- PMID: 13130138
- PMCID: PMC1370488
- DOI: 10.1261/rna.5113603
Efficiency of a programmed -1 ribosomal frameshift in the different subtypes of the human immunodeficiency virus type 1 group M
Abstract
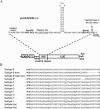
The synthesis of the Gag-Pol polyprotein, the precursor of the enzymes of the human immunodeficiency virus type 1 (HIV-1), requires a programmed -1 ribosomal frameshift. This frameshift has been investigated so far only for subtype B of HIV-1 group M. In this subtype, the frameshift stimulatory signal was found to be a two-stem helix, in which a three-purine bulge interrupts the two stems. In this study, using a luciferase reporter system, we compare, for the first time, the frameshift efficiency of all the subtypes of group M. Mutants of subtype B, including a natural variant were also investigated. Our results with mutants of subtype B confirm that the bulge and the lower stem of the frameshift stimulatory signal contribute to the frameshift in addition to the upper stem-loop considered previously as the sole participant. Our results also show that the frameshift stimulatory signal of all of the other subtypes of group M can be folded into the same structure as in subtype B, despite sequence variations. Moreover, the frameshift efficiency of these subtypes, when assessed in cultured cells, falls within a narrow window (the maximal deviation from the mean value calculated from the experimental values of all the subtypes being approximately 35%), although the predicted thermodynamic stability of the frameshift stimulatory signal differs between the subtypes (from -17.2 kcal/mole to -26.2 kcal/mole). The fact that the frameshift efficiencies fall within a narrow range for all of the subtypes of HIV-1 group M stresses the potential of the frameshift event as an antiviral target.
Figures



Similar articles
-
Characterization of the frameshift stimulatory signal controlling a programmed -1 ribosomal frameshift in the human immunodeficiency virus type 1.Nucleic Acids Res. 2002 Dec 1;30(23):5094-102. doi: 10.1093/nar/gkf657. Nucleic Acids Res. 2002. PMID: 12466532 Free PMC article.
-
Stability of HIV Frameshift Site RNA Correlates with Frameshift Efficiency and Decreased Virus Infectivity.J Virol. 2016 Jul 11;90(15):6906-6917. doi: 10.1128/JVI.00149-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194769 Free PMC article.
-
The virion-associated Gag-Pol is decreased in chimeric Moloney murine leukemia viruses in which the readthrough region is replaced by the frameshift region of the human immunodeficiency virus type 1.Virology. 2005 Apr 10;334(2):342-52. doi: 10.1016/j.virol.2005.01.044. Virology. 2005. PMID: 15780884
-
A review on architecture of the gag-pol ribosomal frameshifting RNA in human immunodeficiency virus: a variability survey of virus genotypes.J Biomol Struct Dyn. 2017 Jun;35(8):1629-1653. doi: 10.1080/07391102.2016.1194231. Epub 2016 Aug 2. J Biomol Struct Dyn. 2017. PMID: 27485859 Review.
-
[The structure of the frameshift stimulatory signal in HIV-1 RNA: a potential target for the treatment of patients infected with HIV].Med Sci (Paris). 2006 Nov;22(11):969-72. doi: 10.1051/medsci/20062211969. Med Sci (Paris). 2006. PMID: 17101099 Review. French.
Cited by
-
Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV.Virus Res. 2006 Jul;119(1):29-42. doi: 10.1016/j.virusres.2005.10.008. Epub 2005 Nov 28. Virus Res. 2006. PMID: 16310880 Free PMC article. Review.
-
The translational landscape of SARS-CoV-2 and infected cells.bioRxiv [Preprint]. 2021 Oct 7:2020.11.03.367516. doi: 10.1101/2020.11.03.367516. bioRxiv. 2021. Update in: mBio. 2022 Jun 28;13(3):e0081522. doi: 10.1128/mbio.00815-22. PMID: 33173862 Free PMC article. Updated. Preprint.
-
High-throughput interrogation of programmed ribosomal frameshifting in human cells.Nat Commun. 2020 Jun 16;11(1):3061. doi: 10.1038/s41467-020-16961-8. Nat Commun. 2020. PMID: 32546731 Free PMC article.
-
HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome.Nucleic Acids Res. 2013 Feb 1;41(3):1901-13. doi: 10.1093/nar/gks1254. Epub 2012 Dec 16. Nucleic Acids Res. 2013. PMID: 23248007 Free PMC article.
-
Mechanisms and biomedical implications of -1 programmed ribosome frameshifting on viral and bacterial mRNAs.FEBS Lett. 2019 Jul;593(13):1468-1482. doi: 10.1002/1873-3468.13478. Epub 2019 Jun 20. FEBS Lett. 2019. PMID: 31222875 Free PMC article. Review.
References
-
- Brierley, I. 1995. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 76: 1885–1892. - PubMed
-
- Brierley, I. and Pennel, S. 2001. Structure and function of the stimulatory RNAs involved in programmed eukaryotic -1 ribosomal frameshifting. In The ribosome, pp 233–248. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
