B-cell and plasma-cell splicing differences: a potential role in regulated immunoglobulin RNA processing
- PMID: 13130140
- PMCID: PMC1370490
- DOI: 10.1261/rna.5820103
B-cell and plasma-cell splicing differences: a potential role in regulated immunoglobulin RNA processing
Abstract
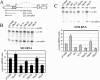
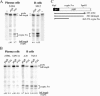
The immunoglobulin micro pre-mRNA is alternatively processed at its 3' end by competing splice and cleavage-polyadenylation reactions to generate mRNAs encoding the membrane-associated or secreted forms of the IgM protein, respectively. The relative use of the competing processing pathways varies during B-lymphocyte development, and it has been established previously that cleavage-polyadenylation activity is higher in plasma cells, which secrete IgM, than in B cells, which produce membrane-associated IgM. To determine whether RNA-splicing activity varies during B-lymphocyte development to contribute to micro RNA-processing regulation, we first demonstrate that micro pre-mRNA processing is sensitive to artificial changes in the splice environment by coexpressing SR proteins with the micro gene. To explore differences between the splice environments of B cells and plasma cells, we analyzed the splicing patterns from two different chimeric non-Ig genes that can be alternatively spliced but have no competing cleavage-polyadenylation reaction. The ratio of intact exon splicing to cryptic splice site use from one chimeric gene differs between several B-cell and several plasma-cell lines. Also, the amount of spliced RNA is higher in B-cell than plasma-cell lines from a set of genes whose splicing is dependent on a functional exonic splice enhancer. Thus, there is clear difference between the B-cell and plasma-cell splicing environments. We propose that both general cleavage-polyadenylation and general splice activities are modulated during B-lymphocyte development to ensure proper regulation of the alternative micro RNA processing pathways.
Figures


Similar articles
-
Alternative processing of IgA pre-mRNA responds like IgM to alterations in the efficiency of the competing splice and cleavage-polyadenylation reactions.Mol Immunol. 1995 Mar;32(4):277-85. doi: 10.1016/0161-5890(94)00141-m. Mol Immunol. 1995. PMID: 7723773
-
Regulated immunoglobulin (Ig) RNA processing does not require specific cis-acting sequences: non-Ig RNA can be alternatively processed in B cells and plasma cells.Mol Cell Biol. 1994 Dec;14(12):7891-8. doi: 10.1128/mcb.14.12.7891-7898.1994. Mol Cell Biol. 1994. PMID: 7969129 Free PMC article.
-
Balanced efficiencies of splicing and cleavage-polyadenylation are required for mu-s and mu-m mRNA regulation.Gene Expr. 1992;2(4):319-27. Gene Expr. 1992. PMID: 1361868 Free PMC article.
-
Immunoglobulin heavy chain gene regulation through polyadenylation and splicing competition.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):92-105. doi: 10.1002/wrna.36. Wiley Interdiscip Rev RNA. 2011. PMID: 21956971 Review.
-
A history of poly A sequences: from formation to factors to function.Prog Nucleic Acid Res Mol Biol. 2002;71:285-389. doi: 10.1016/s0079-6603(02)71046-5. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102557 Review.
Cited by
-
A novel gene expression signature-based on B-cell proportion to predict prognosis of patients with lung adenocarcinoma.BMC Cancer. 2021 Oct 12;21(1):1098. doi: 10.1186/s12885-021-08805-5. BMC Cancer. 2021. PMID: 34641822 Free PMC article.
-
Control of the papillomavirus early-to-late switch by differentially expressed SRp20.J Virol. 2009 Jan;83(1):167-80. doi: 10.1128/JVI.01719-08. Epub 2008 Oct 22. J Virol. 2009. PMID: 18945760 Free PMC article.
-
Cytoplasmic poly(A)-binding protein 1 (PABPC1) interacts with the RNA-binding protein hnRNPLL and thereby regulates immunoglobulin secretion in plasma cells.J Biol Chem. 2017 Jul 21;292(29):12285-12295. doi: 10.1074/jbc.M117.794834. Epub 2017 Jun 13. J Biol Chem. 2017. PMID: 28611064 Free PMC article.
-
A Rapid Method to Characterize Mouse IgG Antibodies and Isolate Native Antigen Binding IgG B Cell Hybridomas.PLoS One. 2015 Aug 28;10(8):e0136613. doi: 10.1371/journal.pone.0136613. eCollection 2015. PLoS One. 2015. PMID: 26317987 Free PMC article.
-
Characterization of a new ARID family transcription factor (Brightlike/ARID3C) that co-activates Bright/ARID3A-mediated immunoglobulin gene transcription.Mol Immunol. 2011 Oct;49(1-2):260-72. doi: 10.1016/j.molimm.2011.08.025. Epub 2011 Sep 28. Mol Immunol. 2011. PMID: 21955986 Free PMC article.
References
-
- Caceres, J.F., Stamm, S., Helfman, D.M., and Krainer, A.R. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265: 1706–1709. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
