A microRNA array reveals extensive regulation of microRNAs during brain development
- PMID: 13130141
- PMCID: PMC1370491
- DOI: 10.1261/rna.5980303
A microRNA array reveals extensive regulation of microRNAs during brain development
Erratum in
- RNA. 2004 Mar;10(3):551
Abstract
Several hundred microRNAs (miRNAs) have recently been cloned from a wide range of organisms across phylogeny. Despite the high degree of conservation of miRNAs, their functions in general, and in mammals particularly, are just beginning to be defined. Here we show that an oligonucleotide DNA array can be successfully used for the simultaneous analysis of miRNA expression profiles from tissues or cells. From a subset of miRNAs expressed in the brain we designed an oligonucleotide array spotted with probes specific for 44 mature miRNAs. These arrays demonstrated precise regulation of miRNA expression at mammalian brain developmental epochs. About 20% of the probed miRNAs changed significantly in their expression during normal brain development, and two of them, miR-9 and miR-131, were dysregulated in presenilin-1 null mice exhibiting severe brain developmental defects. Transcripts with regulated expression patterns on the arrays were validated by Northern blots. Additionally, a bioinformatic analysis of developmentally regulated miRNAs suggested potential mRNA targets. The arrays also revealed miRNAs distributed to translating polyribosomes in primary neurons where they are likely to modulate translation. Therefore, oligonucleotide arrays provide a new tool for studying miRNA expression in a variety of biological and pathobiological settings. Creating clusters of coexpressed miRNAs will contribute to understanding their regulation, functions, and discovery of mRNA targets.
Figures


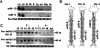



References
-
- Ambros, V. 2000. Control of developmental timing in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 10: 428–433. - PubMed
-
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. 2003. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807–818. - PubMed
-
- Baulcombe, D. 2002. DNA events. An RNA microcosm. Science 297: 2002–2003. - PubMed
-
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
